Twist response of actin filaments
- PMID: 36656858
- PMCID: PMC9942836
- DOI: 10.1073/pnas.2208536120
Twist response of actin filaments
Abstract
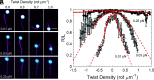
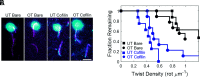
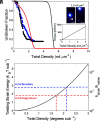
Actin cytoskeleton force generation, sensing, and adaptation are dictated by the bending and twisting mechanics of filaments. Here, we use magnetic tweezers and microfluidics to twist and pull individual actin filaments and evaluate their response to applied loads. Twisted filaments bend and dissipate torsional strain by adopting a supercoiled plectoneme. Pulling prevents plectoneme formation, which causes twisted filaments to sever. Analysis over a range of twisting and pulling forces and direct visualization of filament and single subunit twisting fluctuations yield an actin filament torsional persistence length of ~10 µm, similar to the bending persistence length. Filament severing by cofilin is driven by local twist strain at boundaries between bare and decorated segments and is accelerated by low pN pulling forces. This work explains how contractile forces generated by myosin motors accelerate filament severing by cofilin and establishes a role for filament twisting in the regulation of actin filament stability and assembly dynamics.
Keywords: actin; cofilin; plectoneme; severing; torsion.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- Janmey P. A., McCulloch C. A., Cell mechanics: Integrating cell responses to mechanical stimuli. Annu. Rev. Biomed Eng. 9, 1–34 (2007). - PubMed
-
- Schoen I., Pruitt B. L., Vogel V., The Yin-Yang of rigidity sensing: How forces and mechanical properties regulate the cellular response to materials. Annu. Rev. Mater Res. 43, 589–618 (2013).
-
- Howard J., Mechanics of Motor Proteins and the Cytoskeleton (Sinauer Associates, Publishers, Sunderland, Mass., 2001), p. xvi, 367 p.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

