BTG1 mutation yields supercompetitive B cells primed for malignant transformation
- PMID: 36656933
- PMCID: PMC10515739
- DOI: 10.1126/science.abj7412
BTG1 mutation yields supercompetitive B cells primed for malignant transformation
Abstract
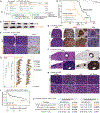
Multicellular life requires altruistic cooperation between cells. The adaptive immune system is a notable exception, wherein germinal center B cells compete vigorously for limiting positive selection signals. Studying primary human lymphomas and developing new mouse models, we found that mutations affecting BTG1 disrupt a critical immune gatekeeper mechanism that strictly limits B cell fitness during antibody affinity maturation. This mechanism converted germinal center B cells into supercompetitors that rapidly outstrip their normal counterparts. This effect was conferred by a small shift in MYC protein induction kinetics but resulted in aggressive invasive lymphomas, which in humans are linked to dire clinical outcomes. Our findings reveal a delicate evolutionary trade-off between natural selection of B cells to provide immunity and potentially dangerous features that recall the more competitive nature of unicellular organisms.
Conflict of interest statement
The authors declare no direct competing financial or nonfinancial interests. K.B.H. receives consulting fees from Prellis Biologics. A.C. is on the immunohistochemistry advisory board for Leica Biosystems and consults for Boehringer Ingelheim Pharmaceuticals. O.E. is supported by Janssen, Johnson and Johnson, Volastra Therapeutics, AstraZeneca, and Eli Lilly research grants; is a scientific adviser to and equity holder in Freenome, Owkin, Volastra Therapeutics, and OneThree Biotech; and consults for and advises Champions Oncology. M.G.K. discloses provision of services with 28-7 Therapeutics (uncompensated), Accent Therapeutics, AstraZeneca, and Kumquat Biosciences; and is a scientific adviser to and equity holder in 858 Therapeutics. S.R.J. is scientific founder of, adviser to, and owns equity in Gotham Therapeutics and 858 Therapeutics. D.W.S. receives research funding from Janssen and NanoString; has consulted for Abbvie, AstraZeneca, Celgene, and Janssen; and was named inventor on patents describing the use of gene expression in subtyping lymphomas. G.D.V. is a scientific adviser for Vaccine Company, Inc. A.M. receives research funding from Janssen Pharmaceuticals, Sanofi, Epizyme, and Daiichi Sankyo; has consulted for Epizyme and Constellation; and is on the advisory board of KDAC Pharma.
Figures



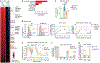

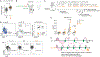
References
MeSH terms
Substances
Grants and funding
- R35 CA220499/CA/NCI NIH HHS/United States
- T32 CA062948/CA/NCI NIH HHS/United States
- K99 CA246080/CA/NCI NIH HHS/United States
- R01 CA186702/CA/NCI NIH HHS/United States
- K99 CA212276/CA/NCI NIH HHS/United States
- R01 AI104739/AI/NIAID NIH HHS/United States
- R01 CA194547/CA/NCI NIH HHS/United States
- F31 CA220981/CA/NCI NIH HHS/United States
- UL1 TR002384/TR/NCATS NIH HHS/United States
- F31 CA254302/CA/NCI NIH HHS/United States
- R01 DK101989/DK/NIDDK NIH HHS/United States
- R01 AI139117/AI/NIAID NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- F31 CA257204/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

