Development of a versatile nuclease prime editor with upgraded precision
- PMID: 36658146
- PMCID: PMC9852468
- DOI: 10.1038/s41467-023-35870-0
Development of a versatile nuclease prime editor with upgraded precision
Abstract
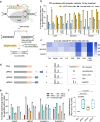
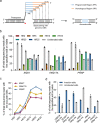
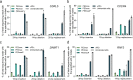
The applicability of nuclease-based form of prime editor (PEn) has been hindered by its complexed editing outcomes. A chemical inhibitor against DNA-PK, which mediates the nonhomologous end joining (NHEJ) pathway, was recently shown to promote precise insertions by PEn. Nevertheless, the intrinsic issues of specificity and toxicity for such a chemical approach necessitate development of alternative strategies. Here, we find that co-introduction of PEn and a NHEJ-restraining, 53BP1-inhibitory ubiquitin variant potently drives precise edits via mitigation of unintended edits, framing a high-activity editing platform (uPEn) apparently complementing the canonical PE. Further developments involve exploring the effective configuration of a homologous region-containing pegRNA (HR-pegRNA). Overall, uPEn can empower high-efficiency installation of insertions (38%), deletions (43%) and replacements (52%) in HEK293T cells. When compared with PE3/5max, uPEn demonstrates superior activities for typically refractory base substitutions, and for small-block edits. Collectively, this work establishes a highly efficient PE platform with broad application potential.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Configuration of adaptable template RNA architectures to unfold the editable space of a nuclease prime editor.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf522. doi: 10.1093/nar/gkaf522. Nucleic Acids Res. 2025. PMID: 40498069 Free PMC article.
-
Harnessing DSB repair to promote efficient homology-dependent and -independent prime editing.Nat Commun. 2022 Mar 24;13(1):1240. doi: 10.1038/s41467-022-28771-1. Nat Commun. 2022. PMID: 35332138 Free PMC article.
-
Dual inhibition of DNA-PK and Polϴ boosts precision of diverse prime editing systems.Nat Commun. 2025 May 8;16(1):4290. doi: 10.1038/s41467-025-59708-z. Nat Commun. 2025. PMID: 40341582 Free PMC article.
-
Designing and executing prime editing experiments in mammalian cells.Nat Protoc. 2022 Nov;17(11):2431-2468. doi: 10.1038/s41596-022-00724-4. Epub 2022 Aug 8. Nat Protoc. 2022. PMID: 35941224 Free PMC article. Review.
-
CRISPR/Cas-based precision genome editing via microhomology-mediated end joining.Plant Biotechnol J. 2021 Feb;19(2):230-239. doi: 10.1111/pbi.13490. Epub 2020 Nov 9. Plant Biotechnol J. 2021. PMID: 33047464 Free PMC article. Review.
Cited by
-
Antiretrovirals to CCR5 CRISPR/Cas9 gene editing - A paradigm shift chasing an HIV cure.Clin Immunol. 2023 Oct;255:109741. doi: 10.1016/j.clim.2023.109741. Epub 2023 Aug 21. Clin Immunol. 2023. PMID: 37611838 Free PMC article. Review.
-
An upgraded nuclease prime editor platform enables high-efficiency singled or multiplexed knock-in/knockout of genes in mouse and sheep zygotes.Protein Cell. 2025 Aug 7;16(8):732-738. doi: 10.1093/procel/pwaf006. Protein Cell. 2025. PMID: 39832212 Free PMC article. No abstract available.
-
Efficient in situ epitope tagging of rice genes by nuclease-mediated prime editing.Plant Cell. 2025 Feb 13;37(2):koae316. doi: 10.1093/plcell/koae316. Plant Cell. 2025. PMID: 39657918
-
Configuration of adaptable template RNA architectures to unfold the editable space of a nuclease prime editor.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf522. doi: 10.1093/nar/gkaf522. Nucleic Acids Res. 2025. PMID: 40498069 Free PMC article.
-
Modified pegRNAs mitigate scaffold-derived prime editing by-products.Nat Commun. 2025 Apr 9;16(1):3374. doi: 10.1038/s41467-025-58653-1. Nat Commun. 2025. PMID: 40204736 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

