scm6A-seq reveals single-cell landscapes of the dynamic m6A during oocyte maturation and early embryonic development
- PMID: 36658155
- PMCID: PMC9852475
- DOI: 10.1038/s41467-023-35958-7
scm6A-seq reveals single-cell landscapes of the dynamic m6A during oocyte maturation and early embryonic development
Abstract
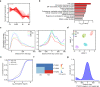
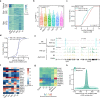
N6-methyladenosine (m6A) has been demonstrated to regulate RNA metabolism and various biological processes, including gametogenesis and embryogenesis. However, the landscape and function of m6A at single cell resolution have not been extensively studied in mammalian oocytes or during pre-implantation. In this study, we developed a single-cell m6A sequencing (scm6A-seq) method to simultaneously profile the m6A methylome and transcriptome in single oocytes/blastomeres of cleavage-stage embryos. We found that m6A deficiency leads to aberrant RNA clearance and consequent low quality of Mettl3Gdf9 conditional knockout (cKO) oocytes. We further revealed that m6A regulates the translation and stability of modified RNAs in metaphase II (MII) oocytes and during oocyte-to-embryo transition, respectively. Moreover, we observed m6A-dependent asymmetries in the epi-transcriptome between the blastomeres of two-cell embryo. scm6A-seq thus allows in-depth investigation into m6A characteristics and functions, and the findings provide invaluable single-cell resolution resources for delineating the underlying mechanism for gametogenesis and early embryonic development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

