Mitochondria regulate intracellular coenzyme Q transport and ferroptotic resistance via STARD7
- PMID: 36658222
- PMCID: PMC9928583
- DOI: 10.1038/s41556-022-01071-y
Mitochondria regulate intracellular coenzyme Q transport and ferroptotic resistance via STARD7
Abstract
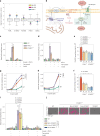
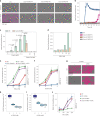
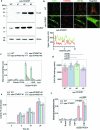
Coenzyme Q (or ubiquinone) is a redox-active lipid that serves as universal electron carrier in the mitochondrial respiratory chain and antioxidant in the plasma membrane limiting lipid peroxidation and ferroptosis. Mechanisms allowing cellular coenzyme Q distribution after synthesis within mitochondria are not understood. Here we identify the cytosolic lipid transfer protein STARD7 as a critical factor of intracellular coenzyme Q transport and suppressor of ferroptosis. Dual localization of STARD7 to the intermembrane space of mitochondria and the cytosol upon cleavage by the rhomboid protease PARL ensures the synthesis of coenzyme Q in mitochondria and its transport to the plasma membrane. While mitochondrial STARD7 preserves coenzyme Q synthesis, oxidative phosphorylation function and cristae morphogenesis, cytosolic STARD7 is required for the transport of coenzyme Q to the plasma membrane and protects against ferroptosis. A coenzyme Q variant competes with phosphatidylcholine for binding to purified STARD7 in vitro. Overexpression of cytosolic STARD7 increases ferroptotic resistance of the cells, but limits coenzyme Q abundance in mitochondria and respiratory cell growth. Our findings thus demonstrate the need to coordinate coenzyme Q synthesis and cellular distribution by PARL-mediated STARD7 processing and identify PARL and STARD7 as promising targets to interfere with ferroptosis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








Comment in
-
START smuggling CoQ to fight ferroptosis.Nat Cell Biol. 2023 Feb;25(2):207-208. doi: 10.1038/s41556-022-01044-1. Nat Cell Biol. 2023. PMID: 36658221 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

