A guide to naming eukaryotic circular RNAs
- PMID: 36658223
- PMCID: PMC10114414
- DOI: 10.1038/s41556-022-01066-9
A guide to naming eukaryotic circular RNAs
Abstract
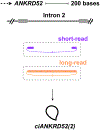
Alternative splicing of eukaryotic transcripts often leads to production of multiple mature RNAs from a single gene locus. In addition to encoding linear RNAs, genes can produce stable circular RNAs (circRNAs) that are often co-expressed with their cognate linear RNAs. Multiple distinct circRNAs are frequently generated from a gene locus via back-splicing, with each mature transcript having a potentially unique function due to its distinct combination of exons and sometimes retained introns. However, names currently given to circRNAs are often ambiguous and lack consistency across studies. Here, we call on the community to embrace standards for naming circRNAs so that a common nomenclature is used to ensure clarity and reproducibility.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

