Structures of the sulfite detoxifying F420-dependent enzyme from Methanococcales
- PMID: 36658338
- PMCID: PMC10229431
- DOI: 10.1038/s41589-022-01232-y
Structures of the sulfite detoxifying F420-dependent enzyme from Methanococcales
Abstract
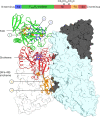
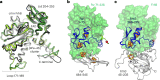
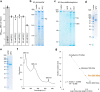
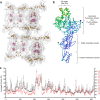
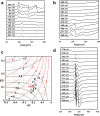
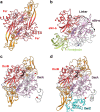
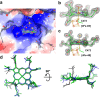
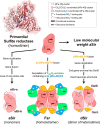
Methanogenic archaea are main actors in the carbon cycle but are sensitive to reactive sulfite. Some methanogens use a sulfite detoxification system that combines an F420H2-oxidase with a sulfite reductase, both of which are proposed precursors of modern enzymes. Here, we present snapshots of this coupled system, named coenzyme F420-dependent sulfite reductase (Group I Fsr), obtained from two marine methanogens. Fsr organizes as a homotetramer, harboring an intertwined six-[4Fe-4S] cluster relay characterized by spectroscopy. The wire, spanning 5.4 nm, electronically connects the flavin to the siroheme center. Despite a structural architecture similar to dissimilatory sulfite reductases, Fsr shows a siroheme coordination and a reaction mechanism identical to assimilatory sulfite reductases. Accordingly, the reaction of Fsr is unidirectional, reducing sulfite or nitrite with F420H2. Our results provide structural insights into this unique fusion, in which a primitive sulfite reductase turns a poison into an elementary block of life.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Jones WJ, Leigh JA, Mayer F, Woese CR, Wolfe RS. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 1983;136:254–261. doi: 10.1007/BF00425213. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

