Engineered live bacteria suppress Pseudomonas aeruginosa infection in mouse lung and dissolve endotracheal-tube biofilms
- PMID: 36658340
- PMCID: PMC10421741
- DOI: 10.1038/s41587-022-01584-9
Engineered live bacteria suppress Pseudomonas aeruginosa infection in mouse lung and dissolve endotracheal-tube biofilms
Abstract
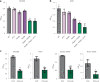
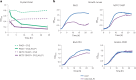
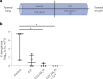
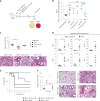
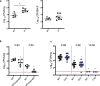
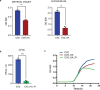
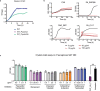
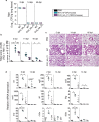
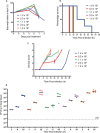
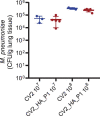
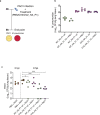
Engineered live bacteria could provide a new modality for treating lung infections, a major cause of mortality worldwide. In the present study, we engineered a genome-reduced human lung bacterium, Mycoplasma pneumoniae, to treat ventilator-associated pneumonia, a disease with high hospital mortality when associated with Pseudomonas aeruginosa biofilms. After validating the biosafety of an attenuated M. pneumoniae chassis in mice, we introduced four transgenes into the chromosome by transposition to implement bactericidal and biofilm degradation activities. We show that this engineered strain has high efficacy against an acute P. aeruginosa lung infection in a mouse model. In addition, we demonstrated that the engineered strain could dissolve biofilms formed in endotracheal tubes of patients with ventilator-associated pneumonia and be combined with antibiotics targeting the peptidoglycan layer to increase efficacy against Gram-positive and Gram-negative bacteria. We expect our M. pneumoniae-engineered strain to be able to treat biofilm-associated infections in the respiratory tract.
© 2023. The Author(s).
Conflict of interest statement
The results published in this article are covered by patents US10745450B2, EP3262061A1 and PCT/EP2021/057122 (licensed to Pulmobiotics S.L) and PCT/EP2021/059142. L.S. and M.L. are shareholders of Pulmobiotics S.L.. R.M., C.P. and M.L. are employees and have stock options of Pulmobiotics S.L. The remaining authors declare no competing interests.
Figures


Comment in
-
Engineered Mycoplasma fight lung infections.Nat Rev Microbiol. 2023 Apr;21(4):218. doi: 10.1038/s41579-023-00863-9. Nat Rev Microbiol. 2023. PMID: 36732447 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

