Whole genome sequencing provides comprehensive genetic testing in childhood B-cell acute lymphoblastic leukaemia
- PMID: 36658389
- PMCID: PMC9991920
- DOI: 10.1038/s41375-022-01806-8
Whole genome sequencing provides comprehensive genetic testing in childhood B-cell acute lymphoblastic leukaemia
Abstract
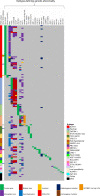
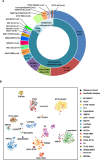
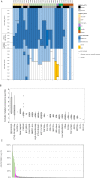
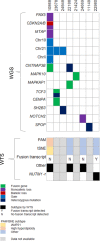
Childhood B-cell acute lymphoblastic leukaemia (B-ALL) is characterised by recurrent genetic abnormalities that drive risk-directed treatment strategies. Using current techniques, accurate detection of such aberrations can be challenging, due to the rapidly expanding list of key genetic abnormalities. Whole genome sequencing (WGS) has the potential to improve genetic testing, but requires comprehensive validation. We performed WGS on 210 childhood B-ALL samples annotated with clinical and genetic data. We devised a molecular classification system to subtype these patients based on identification of key genetic changes in tumour-normal and tumour-only analyses. This approach detected 294 subtype-defining genetic abnormalities in 96% (202/210) patients. Novel genetic variants, including fusions involving genes in the MAP kinase pathway, were identified. WGS results were concordant with standard-of-care methods and whole transcriptome sequencing (WTS). We expanded the catalogue of genetic profiles that reliably classify PAX5alt and ETV6::RUNX1-like subtypes. Our novel bioinformatic pipeline improved detection of DUX4 rearrangements (DUX4-r): a good-risk B-ALL subtype with high survival rates. Overall, we have validated that WGS provides a standalone, reliable genetic test to detect all subtype-defining genetic abnormalities in B-ALL, accurately classifying patients for the risk-directed treatment stratification, while simultaneously performing as a research tool to identify novel disease biomarkers.
© 2023. The Author(s).
Conflict of interest statement
MTR, DRB, JFP, ZK, MM, JB and TJ are employees of Illumina, a public company that develops and markets systems for genetic analysis. CGM has received consulting and advisory board fees from Illumina Inc. and Amgen, and research funding form Pfizer and AbbVie.
Figures
References
-
- Schwab CJ, Murdy D, Butler E, Enshaei A, Winterman E, Cranston RE, et al. Genetic characterisation of childhood B-other-acute lymphoblastic leukaemia in UK patients by fluorescence in situ hybridisation and Multiplex Ligation-dependent Probe Amplification. Br J Haematol. 2021. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

