Genomic autopsy to identify underlying causes of pregnancy loss and perinatal death
- PMID: 36658419
- PMCID: PMC10333122
- DOI: 10.1038/s41591-022-02142-1
Genomic autopsy to identify underlying causes of pregnancy loss and perinatal death
Erratum in
-
Author Correction: Genomic autopsy to identify underlying causes of pregnancy loss and perinatal death.Nat Med. 2024 Jan;30(1):302. doi: 10.1038/s41591-023-02487-1. Nat Med. 2024. PMID: 37429925 No abstract available.
Abstract
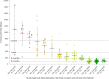
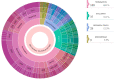
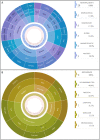
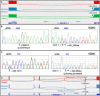
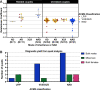
Pregnancy loss and perinatal death are devastating events for families. We assessed 'genomic autopsy' as an adjunct to standard autopsy for 200 families who had experienced fetal or newborn death, providing a definitive or candidate genetic diagnosis in 105 families. Our cohort provides evidence of severe atypical in utero presentations of known genetic disorders and identifies novel phenotypes and disease genes. Inheritance of 42% of definitive diagnoses were either autosomal recessive (30.8%), X-linked recessive (3.8%) or autosomal dominant (excluding de novos, 7.7%), with risk of recurrence in future pregnancies. We report that at least ten families (5%) used their diagnosis for preimplantation (5) or prenatal diagnosis (5) of 12 pregnancies. We emphasize the clinical importance of genomic investigations of pregnancy loss and perinatal death, with short turnaround times for diagnostic reporting and followed by systematic research follow-up investigations. This approach has the potential to enable accurate counseling for future pregnancies.
© 2023. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
Genomic autopsy offers answers for pregnancy loss and perinatal death.Nat Med. 2023 Jan;29(1):41-42. doi: 10.1038/s41591-022-02143-0. Nat Med. 2023. PMID: 36670297 No abstract available.
References
-
- AIHW. Stillbirths and Neonatal Deaths in Australia. https://www.aihw.gov.au/reports/mothers-babies/stillbirths-and-neonatal-... (2020).
-
- Hug L, Alexander M, You D, Alkema L, Inter-agency UN. Group for Child Mortality Estimation. National, regional, and global levels and trends in neonatal mortality between 1990 and 2017, with scenario-based projections to 2030: a systematic analysis. Lancet Glob. Health. 2019;7:e710–e720. doi: 10.1016/S2214-109X(19)30163-9. - DOI - PMC - PubMed
-
- Hoyert, D. L. & Gregory, C. W. Cause of Fetal Death: Data From the Fetal Death Report, 2014. https://www.cdc.gov/nchs/data/nvsr/nvsr65/nvsr65_07.pdf (2016). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

