Hypomethylation in MTNR1B: a novel epigenetic marker for atherosclerosis profiling using stenosis radiophenotype and blood inflammatory cells
- PMID: 36658621
- PMCID: PMC9854223
- DOI: 10.1186/s13148-023-01423-x
Hypomethylation in MTNR1B: a novel epigenetic marker for atherosclerosis profiling using stenosis radiophenotype and blood inflammatory cells
Abstract
Background: Changes in gene-specific promoter methylation may result from aging and environmental influences. Atherosclerosis is associated with aging and environmental effects. Thus, promoter methylation profiling may be used as an epigenetic tool to evaluate the impact of aging and the environment on atherosclerosis development. However, gene-specific methylation changes are currently inadequate epigenetic markers for predicting atherosclerosis and cardiovascular disease pathogenesis.
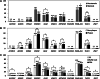
Results: We profiled and validated changes in gene-specific promoter methylation associated with atherosclerosis using stenosis radiophenotypes of cranial vessels and blood inflammatory cells rather than direct sampling of atherosclerotic plaques. First, we profiled gene-specific promoter methylation changes using digital restriction enzyme analysis of methylation (DREAM) sequencing in peripheral blood mononuclear cells from eight samples each of cranial vessels with and without severe-stenosis radiophenotypes. Using DREAM sequencing profiling, 11 tags were detected in the promoter regions of the ACVR1C, ADCK5, EFNA2, ENOSF1, GLS2, KNDC1, MTNR1B, PACSIN3, PAX8-AS1, TLDC1, and ZNF7 genes. Using methylation evaluation, we found that EFNA2, ENOSF1, GLS2, KNDC1, MTNR1B, PAX8-AS1, and TLDC1 showed > 5% promoter methylation in non-plaque intima, atherosclerotic vascular tissues, and buffy coats. Using logistic regression analysis, we identified hypomethylation of MTNR1B as an independent variable for the stenosis radiophenotype prediction model by combining it with traditional atherosclerosis risk factors including age, hypertension history, and increases in creatinine, lipoprotein (a), and homocysteine. We performed fivefold cross-validation of the prediction model using 384 patients with ischemic stroke (50 [13%] no-stenosis and 334 [87%] > 1 stenosis radiophenotype). For the cross-validation, the training dataset included 70% of the dataset. The prediction model showed an accuracy of 0.887, specificity to predict stenosis radiophenotype of 0.940, sensitivity to predict no-stenosis radiophenotype of 0.533, and area under receiver operating characteristic curve of 0.877 to predict stenosis radiophenotype from the test dataset including 30% of the dataset.
Conclusions: We identified and validated MTNR1B hypomethylation as an epigenetic marker to predict cranial vessel atherosclerosis using stenosis radiophenotypes and blood inflammatory cells rather than direct atherosclerotic plaque sampling.
Keywords: Atherosclerosis; Blood inflammatory cells; Promoter methylation profiling; Radioepigenomics; Stenosis radiophenotype.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

