PDZD8-deficient mice manifest behavioral abnormalities related to emotion, cognition, and adaptation due to dyslipidemia in the brain
- PMID: 36658656
- PMCID: PMC9854033
- DOI: 10.1186/s13041-023-01002-4
PDZD8-deficient mice manifest behavioral abnormalities related to emotion, cognition, and adaptation due to dyslipidemia in the brain
Abstract
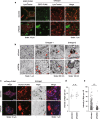
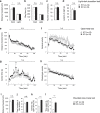
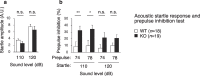
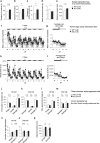
Although dyslipidemia in the brain has been implicated in neurodegenerative disorders, the molecular mechanisms underlying its pathogenesis have been largely unclear. PDZD8 is a lipid transfer protein and mice deficient in PDZD8 (PDZD8-KO mice) manifest abnormal accumulation of cholesteryl esters (CEs) in the brain due to impaired lipophagy, the degradation system of lipid droplets. Here we show the detailed mechanism of PDZD8-dependent lipophagy. PDZD8 transports cholesterol to lipid droplets (LDs), and eventually promotes fusion of LDs and lysosomes. In addition, PDZD8-KO mice exhibit growth retardation, hyperactivity, reduced anxiety and fear, increased sensorimotor gating, and impaired cued fear conditioned memory and working memory. These results indicate that abnormal CE accumulation in the brain caused by PDZD8 deficiency affects emotion, cognition and adaptive behavior, and that PDZD8 plays an important role in the maintenance of brain function through lipid metabolism.
Keywords: Behavior; Cholesterol; Dyslipidemia; Knockout mouse; Lipophagy; PDZD8.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Nugent AA, Lin K, van Lengerich B, Lianoglou S, Przybyla L, Davis SS, et al. TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron. 2020;105(837–54):e9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

