MUG: A mutation overview of GPCR subfamily A17 receptors
- PMID: 36659920
- PMCID: PMC9822836
- DOI: 10.1016/j.csbj.2022.12.031
MUG: A mutation overview of GPCR subfamily A17 receptors
Abstract
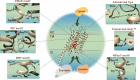
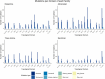
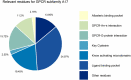
G protein-coupled receptors (GPCRs) mediate several signaling pathways through a general mechanism that involves their activation, upholding a chain of events that lead to the release of molecules responsible for cytoplasmic action and further regulation. These physiological functions can be severely altered by mutations in GPCR genes. GPCRs subfamily A17 (dopamine, serotonin, adrenergic and trace amine receptors) are directly related with neurodegenerative diseases, and as such it is crucial to explore known mutations on these systems and their impact in structure and function. A comprehensive and detailed computational framework - MUG (Mutations Understanding GPCRs) - was constructed, illustrating key reported mutations and their effect on receptors of the subfamily A17 of GPCRs. We explored the type of mutations occurring overall and in the different families of subfamily A17, as well their localization within the receptor and potential effects on receptor functionality. The mutated residues were further analyzed considering their pathogenicity. The results reveal a high diversity of mutations in the GPCR subfamily A17 structures, drawing attention to the considerable number of mutations in conserved residues and domains. Mutated residues were typically hydrophobic residues enriched at the ligand binding pocket and known activating microdomains, which may lead to disruption of receptor function. MUG as an interactive web application is available for the management and visualization of this dataset. We expect that this interactive database helps the exploration of GPCR mutations, their influence, and their familywise and receptor-specific effects, constituting the first step in elucidating their structures and molecules at the atomic level.
Keywords: Database; ECL, extracellular loop; FDA, Food and Drug Administration; G protein-coupled receptors; GEFs, GTP-exchange factors; GOF, gain-of-function; GPCR subfamily A17; GPCRs, G protein-coupled receptors; ICL, intracellular loop; LOF, loss-of-function; MUG, mutations understanding GPCRs; Natural variants; Neurodegenerative diseases; TM, transmembrane.
© 2022 The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures




References
-
- Schöneberg T., Liebscher I. Mutations in G protein-coupled receptors: mechanisms, pathophysiology and potential therapeutic approaches. Pharmacol Rev. 2021;73:89–119. - PubMed
LinkOut - more resources
Full Text Sources
