Mapping the signaling network of BIN2 kinase using TurboID-mediated biotin labeling and phosphoproteomics
- PMID: 36660928
- PMCID: PMC10015162
- DOI: 10.1093/plcell/koad013
Mapping the signaling network of BIN2 kinase using TurboID-mediated biotin labeling and phosphoproteomics
Abstract
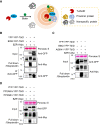
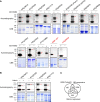
Elucidating enzyme-substrate relationships in posttranslational modification (PTM) networks is crucial for understanding signal transduction pathways but is technically difficult because enzyme-substrate interactions tend to be transient. Here, we demonstrate that TurboID-based proximity labeling (TbPL) effectively and specifically captures the substrates of kinases and phosphatases. TbPL-mass spectrometry (TbPL-MS) identified over 400 proximal proteins of Arabidopsis thaliana BRASSINOSTEROID-INSENSITIVE2 (BIN2), a member of the GLYCOGEN SYNTHASE KINASE 3 (GSK3) family that integrates signaling pathways controlling diverse developmental and acclimation processes. A large portion of the BIN2-proximal proteins showed BIN2-dependent phosphorylation in vivo or in vitro, suggesting that these are BIN2 substrates. Protein-protein interaction network analysis showed that the BIN2-proximal proteins include interactors of BIN2 substrates, revealing a high level of interactions among the BIN2-proximal proteins. Our proteomic analysis establishes the BIN2 signaling network and uncovers BIN2 functions in regulating key cellular processes such as transcription, RNA processing, translation initiation, vesicle trafficking, and cytoskeleton organization. We further discovered significant overlap between the GSK3 phosphorylome and the O-GlcNAcylome, suggesting an evolutionarily ancient relationship between GSK3 and the nutrient-sensing O-glycosylation pathway. Our work presents a powerful method for mapping PTM networks, a large dataset of GSK3 kinase substrates, and important insights into the signaling network that controls key cellular functions underlying plant growth and acclimation.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




Comment in
-
Proximity labeling reveals a BIN2 signaling network.Plant Cell. 2023 Mar 15;35(3):958-959. doi: 10.1093/plcell/koad006. Plant Cell. 2023. PMID: 36650120 Free PMC article. No abstract available.
References
-
- Anne P, Azzopardi M, Gissot L, Beaubiat S, Hématy K, Palauqui JC (2015) OCTOPUS negatively regulates BIN2 to control phloem differentiation in Arabidopsis thaliana. Curr Biol 25(19): 2584–2590 - PubMed
-
- Banerjee PS, Lagerlöf O, Hart GW (2016) Roles of O-GlcNAc in chronic diseases of aging. Mol Aspects Med 51: 1–15 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

