Low HER2 expression in normal breast epithelium enables dedifferentiation and malignant transformation via chromatin opening
- PMID: 36661191
- PMCID: PMC9922733
- DOI: 10.1242/dmm.049894
Low HER2 expression in normal breast epithelium enables dedifferentiation and malignant transformation via chromatin opening
Abstract
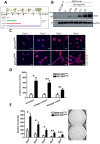
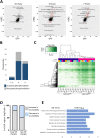
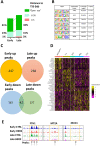
Overexpression of the HER2 protein in breast cancer patients is a predictor of poor prognosis and resistance to therapies. We used an inducible breast cancer transformation system that allows investigation of early molecular changes. HER2 overexpression to similar levels as those observed in a subtype of HER2-positive breast cancer patients induced transformation of MCF10A cells and resulted in gross morphological changes, increased anchorage-independent growth of cells, and altered the transcriptional programme of genes associated with oncogenic transformation. Global phosphoproteomic analysis during HER2 induction predominantly detected an increase in protein phosphorylation. Intriguingly, this correlated with chromatin opening, as measured by ATAC-seq on acini isolated from 3D cell culture. HER2 overexpression resulted in opening of many distal regulatory regions and promoted reprogramming-associated heterogeneity. We found that a subset of cells acquired a dedifferentiated breast stem-like phenotype, making them likely candidates for malignant transformation. Our data show that this population of cells, which counterintuitively enriches for relatively low HER2 protein abundance and increased chromatin accessibility, possesses transformational drive, resulting in increased anchorage-independent growth in vitro compared to cells not displaying a stem-like phenotype.
Keywords: In vitro; Breast; Cancer; Chromatin; Epigenetics; Stem.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

