A chromosome-scale reference genome assembly of the great sand eel, Hyperoplus lanceolatus
- PMID: 36661278
- PMCID: PMC10078159
- DOI: 10.1093/jhered/esad003
A chromosome-scale reference genome assembly of the great sand eel, Hyperoplus lanceolatus
Abstract
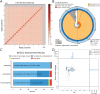
Despite increasing sequencing efforts, numerous fish families still lack a reference genome, which complicates genetic research. One such understudied family is the sand lances (Ammodytidae, literally: "sand burrower"), a globally distributed clade of over 30 fish species that tend to avoid tidal currents by burrowing into the sand. Here, we present the first annotated chromosome-level genome assembly of the great sand eel (Hyperoplus lanceolatus). The genome assembly was generated using Oxford Nanopore Technologies long sequencing reads and Illumina short reads for polishing. The final assembly has a total length of 808.5 Mbp, of which 97.1% were anchored into 24 chromosome-scale scaffolds using proximity-ligation scaffolding. It is highly contiguous with a scaffold and contig N50 of 33.7 and 31.3 Mbp, respectively, and has a BUSCO completeness score of 96.9%. The presented genome assembly is a valuable resource for future studies of sand lances, as this family is of great ecological and commercial importance and may also contribute to studies aiming to resolve the suprafamiliar taxonomy of bony fishes.
Keywords: Ammodytidae; Eupercaria; Omni-C; Oxford Nanopore; proximity-ligation scaffolding.
© The American Genetic Association. 2023.
Conflict of interest statement
None declared.
Figures

References
-
- Dunn E. Revive our Seas: the case for stronger regulation of sandeel fisheries in UK waters. The Royal Society for the Protection of Birds (RSPB); 2021. [accessed 2022 Jun 1]. https://www.rspb.org.uk/globalassets/downloads/documents/campaigning-for...
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

