Structure-based prediction of T cell receptor:peptide-MHC interactions
- PMID: 36661395
- PMCID: PMC9859041
- DOI: 10.7554/eLife.82813
Structure-based prediction of T cell receptor:peptide-MHC interactions
Abstract
The regulatory and effector functions of T cells are initiated by the binding of their cell-surface T cell receptor (TCR) to peptides presented by major histocompatibility complex (MHC) proteins on other cells. The specificity of TCR:peptide-MHC interactions, thus, underlies nearly all adaptive immune responses. Despite intense interest, generalizable predictive models of TCR:peptide-MHC specificity remain out of reach; two key barriers are the diversity of TCR recognition modes and the paucity of training data. Inspired by recent breakthroughs in protein structure prediction achieved by deep neural networks, we evaluated structural modeling as a potential avenue for prediction of TCR epitope specificity. We show that a specialized version of the neural network predictor AlphaFold can generate models of TCR:peptide-MHC interactions that can be used to discriminate correct from incorrect peptide epitopes with substantial accuracy. Although much work remains to be done for these predictions to have widespread practical utility, we are optimistic that deep learning-based structural modeling represents a path to generalizable prediction of TCR:peptide-MHC interaction specificity.
Keywords: MHC; T cell epitope; T cell receptor; binding specificity; computational biology; deep learning; human; immunology; inflammation; mouse; structure prediction; systems biology.
© 2023, Bradley.
Conflict of interest statement
PB No competing interests declared
Figures




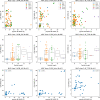





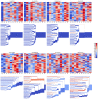






References
-
- 10xGenomics A new way of exploring immunity: linking highly multiplexed antigen recognition to immune repertoire and phenotype. 2020. [June 1, 2021]. https://pages.10xgenomics.com/rs/446-PBO-704/images/10x_AN047_IP_A_New_W...
-
- Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millán C, Park H, Adams C, Glassman CR, DeGiovanni A, Pereira JH, Rodrigues AV, van Dijk AA, Ebrecht AC, Opperman DJ, Sagmeister T, Buhlheller C, Pavkov-Keller T, Rathinaswamy MK, Dalwadi U, Yip CK, Burke JE, Garcia KC, Grishin NV, Adams PD, Read RJ, Baker D. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. - DOI - PMC - PubMed
-
- Beringer DX, Kleijwegt FS, Wiede F, van der Slik AR, Loh KL, Petersen J, Dudek NL, Duinkerken G, Laban S, Joosten A, Vivian JP, Chen Z, Uldrich AP, Godfrey DI, McCluskey J, Price DA, Radford KJ, Purcell AW, Nikolic T, Reid HH, Tiganis T, Roep BO, Rossjohn J. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nature Immunology. 2015;16:1153–1161. doi: 10.1038/ni.3271. - DOI - PubMed
-
- Berkhoff EGM, de Wit E, Geelhoed-Mieras MM, Boon ACM, Symons J, Fouchier RAM, Osterhaus ADME, Rimmelzwaan GF. Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes. Journal of Virology. 2005;79:11239–11246. doi: 10.1128/JVI.79.17.11239-11246.2005. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

