Limited Metabolomic Overlap between Commensal Bacteria and Marine Sponge Holobionts Revealed by Large Scale Culturing and Mass Spectrometry-Based Metabolomics: An Undergraduate Laboratory Pedagogical Effort at Georgia Tech
- PMID: 36662226
- PMCID: PMC9862627
- DOI: 10.3390/md21010053
Limited Metabolomic Overlap between Commensal Bacteria and Marine Sponge Holobionts Revealed by Large Scale Culturing and Mass Spectrometry-Based Metabolomics: An Undergraduate Laboratory Pedagogical Effort at Georgia Tech
Abstract
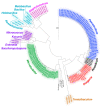
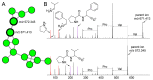
Sponges are the richest source of bioactive organic small molecules, referred to as natural products, in the marine environment. It is well established that laboratory culturing-resistant symbiotic bacteria residing within the eukaryotic sponge host matrix often synthesize the natural products that are detected in the sponge tissue extracts. However, the contributions of the culturing-amenable commensal bacteria that are also associated with the sponge host to the overall metabolome of the sponge holobiont are not well defined. In this study, we cultured a large library of bacteria from three marine sponges commonly found in the Florida Keys. Metabolomes of isolated bacterial strains and that of the sponge holobiont were compared using mass spectrometry to reveal minimal metabolomic overlap between commensal bacteria and the sponge hosts. We also find that the phylogenetic overlap between cultured commensal bacteria and that of the sponge microbiome is minimal. Despite these observations, the commensal bacteria were found to be a rich resource for novel natural product discovery. Mass spectrometry-based metabolomics provided structural insights into these cryptic natural products. Pedagogic innovation in the form of laboratory curricula development is described which provided undergraduate students with hands-on instruction in microbiology and natural product discovery using metabolomic data mining strategies.
Keywords: bacteria; mass spectrometry; metabolomics; natural products; sponge.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- McMurray S.E., Stubler A.D., Erwin P.M., Finelli C.M., Pawlik J.R. A test of the sponge-loop hypothesis for emergent Caribbean reef sponges. Mar. Ecol. Prog. Ser. 2018;588:1–14. doi: 10.3354/meps12466. - DOI
-
- Bart M.C., Hudspith M., Rapp H.T., Verdonschot P.F.M., de Goeij J.M. A deep-sea sponge loop? Sponges transfer dissolved and particulate organic carbon and nitrogen to associated fauna. Front. Mar. Sci. 2021;8:604879. doi: 10.3389/fmars.2021.604879. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

