A chromosome-level reference genome and pangenome for barn swallow population genomics
- PMID: 36662619
- PMCID: PMC10044405
- DOI: 10.1016/j.celrep.2023.111992
A chromosome-level reference genome and pangenome for barn swallow population genomics
Abstract
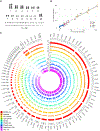
Insights into the evolution of non-model organisms are limited by the lack of reference genomes of high accuracy, completeness, and contiguity. Here, we present a chromosome-level, karyotype-validated reference genome and pangenome for the barn swallow (Hirundo rustica). We complement these resources with a reference-free multialignment of the reference genome with other bird genomes and with the most comprehensive catalog of genetic markers for the barn swallow. We identify potentially conserved and accelerated genes using the multialignment and estimate genome-wide linkage disequilibrium using the catalog. We use the pangenome to infer core and accessory genes and to detect variants using it as a reference. Overall, these resources will foster population genomics studies in the barn swallow, enable detection of candidate genes in comparative genomics studies, and help reduce bias toward a single reference genome.
Keywords: CP: Molecular biology; barn swallow; comparative genomics; genetic marker catalog; genome assembly; linkage disequilibrium; pangenome graph; pangenomics; population genomics; reference genome; synanthropy.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.S. and K.W. are full-time employees at Pacific Biosciences, a company commercializing single-molecule sequencing technologies.
Figures


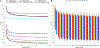

References
-
- Spina F (1998). The EURING swallow project: a large-scale approach to the study and conservation of a long-distance migrant. Migrating birds know no boundaries. Proc. Int. Symp. Isr. Torgos 28, 151–162.
-
- Johnston RF (2001). Synanthropic birds of North America. In Avian Ecology and Conservation in an Urbanizing World, Marzluff JM, Bowman R, and Donnelly R, eds. (Springer US; ), pp. 49–67. 10.1007/978-1-4615-1531-9_3. - DOI
-
- Krajcarz M, Krajcarz MT, Baca M, Baumann C, Van Neer W, Popović D, Sudo1-Procyk M, Wach B, Wilczyński J, Wojenka M, et al. (2020). Ancestors of domestic cats in Neolithic Central Europe: Isotopic evidence of a synanthropic diet. Proc. Natl. Acad. Sci. USA 117, 17710–17719. 10.1073/pnas.1918884117. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

