Origins of genome-editing excisases as illuminated by the somatic genome of the ciliate Blepharisma
- PMID: 36669098
- PMCID: PMC9942806
- DOI: 10.1073/pnas.2213887120
Origins of genome-editing excisases as illuminated by the somatic genome of the ciliate Blepharisma
Abstract
Massive DNA excision occurs regularly in ciliates, ubiquitous microbial eukaryotes with somatic and germline nuclei in the same cell. Tens of thousands of internally eliminated sequences (IESs) scattered throughout the ciliate germline genome are deleted during the development of the streamlined somatic genome. The genus Blepharisma represents one of the two high-level ciliate clades (subphylum Postciliodesmatophora) and, unusually, has dual pathways of somatic nuclear and genome development. This makes it ideal for investigating the functioning and evolution of these processes. Here we report the somatic genome assembly of Blepharisma stoltei strain ATCC 30299 (41 Mbp), arranged as numerous telomere-capped minichromosomal isoforms. This genome encodes eight PiggyBac transposase homologs no longer harbored by transposons. All appear subject to purifying selection, but just one, the putative IES excisase, has a complete catalytic triad. We hypothesize that PiggyBac homologs were ancestral excisases that enabled the evolution of extensive natural genome editing.
Keywords: PiggyBac; PiggyMac; natural genome editing; transposase; transposon.
Conflict of interest statement
The authors declare no competing interest.
Figures

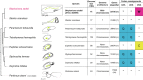
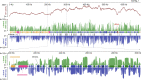
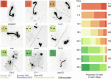

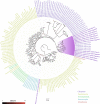
Similar articles
-
Nuclear dualism without extensive DNA elimination in the ciliate Loxodes magnus.Proc Natl Acad Sci U S A. 2024 Sep 24;121(39):e2400503121. doi: 10.1073/pnas.2400503121. Epub 2024 Sep 19. Proc Natl Acad Sci U S A. 2024. PMID: 39298487 Free PMC article.
-
MITE infestation accommodated by genome editing in the germline genome of the ciliate Blepharisma.Proc Natl Acad Sci U S A. 2023 Jan 24;120(4):e2213985120. doi: 10.1073/pnas.2213985120. Epub 2023 Jan 20. Proc Natl Acad Sci U S A. 2023. PMID: 36669106 Free PMC article.
-
Ku-mediated coupling of DNA cleavage and repair during programmed genome rearrangements in the ciliate Paramecium tetraurelia.PLoS Genet. 2014 Aug 28;10(8):e1004552. doi: 10.1371/journal.pgen.1004552. eCollection 2014 Aug. PLoS Genet. 2014. PMID: 25166013 Free PMC article.
-
Programmed Rearrangement in Ciliates: Paramecium.Microbiol Spectr. 2014 Dec;2(6). doi: 10.1128/microbiolspec.MDNA3-0035-2014. Microbiol Spectr. 2014. PMID: 26104450 Review.
-
Transposon domestication versus mutualism in ciliate genome rearrangements.PLoS Genet. 2013;9(8):e1003659. doi: 10.1371/journal.pgen.1003659. Epub 2013 Aug 1. PLoS Genet. 2013. PMID: 23935529 Free PMC article. Review.
Cited by
-
Unveiling an ancient whole-genome duplication event in Stentor, the model unicellular eukaryotes.Sci China Life Sci. 2025 Mar;68(3):825-835. doi: 10.1007/s11427-024-2651-2. Epub 2025 Jan 8. Sci China Life Sci. 2025. PMID: 39821159
-
Nuclear dualism without extensive DNA elimination in the ciliate Loxodes magnus.Proc Natl Acad Sci U S A. 2024 Sep 24;121(39):e2400503121. doi: 10.1073/pnas.2400503121. Epub 2024 Sep 19. Proc Natl Acad Sci U S A. 2024. PMID: 39298487 Free PMC article.
-
The cell biology and genome of Stentor pyriformis, a giant cell that embeds symbiotic algae in a microtubule meshwork.Mol Biol Cell. 2025 Apr 1;36(4):ar44. doi: 10.1091/mbc.E24-12-0571. Epub 2025 Feb 12. Mol Biol Cell. 2025. PMID: 39937680 Free PMC article.
-
Functions and mechanisms of eukaryotic RNA-guided programmed DNA elimination.Biochem Soc Trans. 2025 Apr 29;53(2):473-85. doi: 10.1042/BST20253006. Biochem Soc Trans. 2025. PMID: 40305257 Free PMC article. Review.
-
How ciliates got their nuclei.Proc Natl Acad Sci U S A. 2023 Feb 14;120(7):e2221818120. doi: 10.1073/pnas.2221818120. Epub 2023 Feb 10. Proc Natl Acad Sci U S A. 2023. PMID: 36763531 Free PMC article. No abstract available.
References
-
- Swart E. C., Nowacki M., The eukaryotic way to defend and edit genomes by sRNA-targeted DNA deletion. Ann. N. Y. Acad. Sci. 1341, 106–114 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

