Cell survival following direct executioner-caspase activation
- PMID: 36669100
- PMCID: PMC9942801
- DOI: 10.1073/pnas.2216531120
Cell survival following direct executioner-caspase activation
Erratum in
-
Correction for Nano et al., Cell survival following direct executioner-caspase activation.Proc Natl Acad Sci U S A. 2023 Nov 14;120(46):e2317216120. doi: 10.1073/pnas.2317216120. Epub 2023 Nov 9. Proc Natl Acad Sci U S A. 2023. PMID: 37943839 Free PMC article. No abstract available.
Abstract
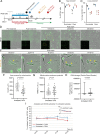
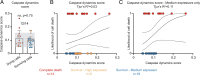
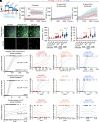
Executioner-caspase activation has been considered a point-of-no-return in apoptosis. However, numerous studies report survival from caspase activation after treatment with drugs or radiation. An open question is whether cells can recover from direct caspase activation without pro-survival stress responses induced by drugs. To address this question, we engineered a HeLa cell line to express caspase-3 inducibly and combined it with a quantitative caspase activity reporter. While high caspase activity levels killed all cells and very low levels allowed all cells to live, doses of caspase activity sufficient to kill 15 to 30% of cells nevertheless allowed 70 to 85% to survive. At these doses, neither the rate, nor the peak level, nor the total amount of caspase activity could accurately predict cell death versus survival. Thus, cells can survive direct executioner-caspase activation, and variations in cellular state modify the outcome of potentially lethal caspase activity. Such heterogeneities may underlie incomplete tumor cell killing in response to apoptosis-inducing cancer treatments.
Keywords: anastasis; apoptosis; effector caspase; predictive power; recovery.
Conflict of interest statement
The authors have organizational affiliations, stock ownership, and research support to disclose. D.J.M. is a member of the Board of Scientific Counselors for the National Cancer Institute; has 5% equity in Mór Bio, a subsidiary of Inceptor Bio, which is a cellular immunotherapy company; and has 20% equity in Anastasis Biotechnology Corporation. The authors have patent filings to disclose: Modulation of Anastasis, UCSB Case Number 2020-062-2, US Patent found in United States Provisional Patent Application Number 63/029,380, entitled “Methods of Modulating Anastasis,” filed May 22, 2020; Detection of Anastasis, UCSB Case Number 2020-62-1, found in US Patent United States Provisional Patent Application Number 63/029,358, entitled “Methods of Detecting Anastasis,” filed May 22, 2020; US provisional application no. 63/014,049 entitled “Stimulating US Patent Phagocytosis of Cancer Cells by Activating Rac in Macrophages” filed on April 23, 2020, with docket number P2495-USP; and US provisional application no. 63/126,379 entitled “Genetically US Patent Engineered Phagocytes and Related Compositions Methods and Systems” filed on December 16, 2020, with docket number P2495-USP2. D.J.M. has received research funds in the form of unrestricted gifts from the Anastasis Biotechnology Corporation and from Inceptor Bio.
Figures



References
-
- Cavallucci V., D’Amelio M., "Physiological and pathological role of apoptosis" in Apoptosome: An up-and-Coming Therapeutical Tool, Cecconi F., D’Amelio M., Eds. (Springer, Netherlands, 2010), pp. 1–26.
-
- Zhang P., Su D.-M., Liang M., Fu J., Chemopreventive agents induce programmed death-1-ligand 1 (PD-L1) surface expression in breast cancer cells and promote PD-L1-mediated T cell apoptosis. Mol. Immunol. 45, 1470–1476 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

