BAP1 Loss Is Associated with Higher ASS1 Expression in Epithelioid Mesothelioma: Implications for Therapeutic Stratification
- PMID: 36669126
- PMCID: PMC10150242
- DOI: 10.1158/1541-7786.MCR-22-0635
BAP1 Loss Is Associated with Higher ASS1 Expression in Epithelioid Mesothelioma: Implications for Therapeutic Stratification
Abstract
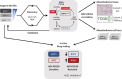
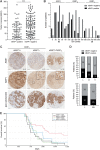
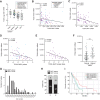
The nuclear deubiquitylase BRCA1-associated protein 1 (BAP1) is frequently inactivated in malignant pleural mesothelioma (MPM) and germline BAP1 mutation predisposes to cancers including MPM. To explore the influence on cell physiology and drug sensitivity, we sequentially edited a predisposition mutation (w-) and a promoter trap (KO) into human mesothelial cells. BAP1w-/KO MeT5A cells express less BAP1 protein and phenocopy key aspects of BAP1 loss in MPM. Stable isotope labeling with amino acids in cell culture-mass spectrometry revealed evidence of metabolic adaptation, with concomitant alteration of cellular metabolites. In MeT5A, BAP1 deficiency reduces glycolytic enzyme levels but increases enzymes involved in the tricarboxylic acid cycle and anaplerotic pathways. Notably both argininosuccinate synthase 1 (ASS1), essential for cellular synthesis of arginine, and its substrate aspartate, are elevated in BAP1w-/KO MeT5A cells. Likewise, ASS1 expression is higher in BAP1-altered MPM cell lines, and inversely correlates with BAP1 in The Cancer Genome Atlas MESO dataset. Elevated ASS1 is also evident by IHC staining in epithelioid MPM lacking nuclear BAP1 expression, with improved survival among patients with BAP1-negative/ASS1-expressing tumors. Alterations in arginine metabolism may sensitize cells to metabolic drugs and we find that BAP1-negative/ASS1-expressing MPM cell lines are more sensitive to ASS1 inhibition, although not to inhibition of purine synthesis by mizoribine. Importantly, BAP1w-/KO MeT5A become desensitized to arginine deprivation by pegylated arginine deiminase (ADI-PEG20), phenocopying BAP1-negative/ASS1-expressing MPM cell lines.
Implications: Our data reveal an interrelationship between BAP1 and arginine metabolism, providing a potential means of identifying patients with epithelioid MPM likely to benefit from ADI-PEG20.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures





Comment in
- 1541-7786. doi: 10.1158/1541-7786.MCR-21-5-HI
References
-
- Takahashi K, Landrigan PJ, Collegium R. The global health dimensions of asbestos and asbestos-related diseases. Ann Glob Health 2016;82:209–13. - PubMed
-
- Zalcman G, Mazieres J, Margery J, Greillier L, Audigier-Valette C, Moro-Sibilot D, et al. . Bevacizumab for newly diagnosed pleural mesothelioma in the mesothelioma avastin cisplatin pemetrexed study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet 2016;387:1405–14. - PubMed
-
- Baas P, Scherpereel A, Nowak AK, Fujimoto N, Peters S, Tsao AS, et al. . First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): a multicentre, randomised, open-label, phase 3 trial. Lancet 2021;397:375–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/R009120/1/MRC_/Medical Research Council/United Kingdom
- NC/T001631/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- 102382/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- G1002610/MRC_/Medical Research Council/United Kingdom
- MR/V028669/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

