Population differentiated copy number variation between Eurasian wild boar and domesticated pig populations
- PMID: 36670113
- PMCID: PMC9859782
- DOI: 10.1038/s41598-022-22373-z
Population differentiated copy number variation between Eurasian wild boar and domesticated pig populations
Abstract
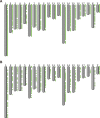
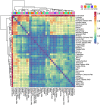
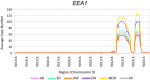
Sus scrofa is a globally distributed livestock species that still maintains two different ways of life: wild and domesticated. Herein, we detected copy number variation (CNV) of 328 animals using short read alignment on Sscrofa11.1. We compared CNV among five groups of porcine populations: Asian domesticated (AD), European domesticated (ED), Asian wild (AW), European wild (EW), and Near Eastern wild (NEW). In total, 21,673 genes were identified on 154,872 copy number variation region (CNVR). Differences in gene copy numbers between populations were measured by considering the variance-based value [Formula: see text] and the one-way ANOVA test followed by Scheffe test. As a result, 111 genes were suggested as copy number variable genes. Abnormally gained copy number on EEA1 in all populations was suggested the presence of minor CNV in the reference genome assembly, Sscrofa11.1. Copy number variable genes were related to meat quality, immune response, and reproduction traits. Hierarchical clustering of all individuals and mean pairwise [Formula: see text] in breed level were visualized genetic relationship of 328 individuals and 56 populations separately. Our findings have shown how the complex history of pig evolution appears in genome-wide CNV of various populations with different regions and lifestyles.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

