The structure and diversity of microbial communities in Paederus fuscipes (Coleoptera: Staphylinidae): from ecological paradigm to pathobiome
- PMID: 36670494
- PMCID: PMC9862579
- DOI: 10.1186/s40168-022-01456-z
The structure and diversity of microbial communities in Paederus fuscipes (Coleoptera: Staphylinidae): from ecological paradigm to pathobiome
Abstract
Background: Paederus fuscipes is medically the most famous rove beetle, which causes dermatitis or conjunctivitis in humans, as well as gastrointestinal toxicosis in livestock, via releasing toxic hemolymph containing pederin. Pedrin biosynthesis genes have been identified in uncultured Pseudomonas-like endosymbionts that are speculated to be acquired through a horizontal transfer. However, the composition of the P. fuscipes microbial community, especially of the gut and genital microbiome, remains unclear. This study was aimed to characterize the structure and diversity of P. fuscipes-associated bacterial communities in terms of gender, organ, and location using the Illumina HiSeq platform in the southern littorals of Caspian Sea.
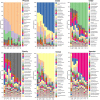
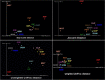
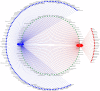
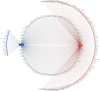
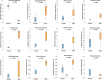
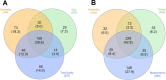
Results: The OTUs identified from P. fuscipes specimens were collapsed into 40 phyla, 112 classes, 249 orders, 365 families, 576 genera, and 106 species. The most abundant families were Pseudomonadaceae, Spiroplasmataceae, Weeksellaceae, Enterococcaceae, and Rhizobiaceae, respectively. Thirty top genera made up > 94% of the P. fuscipes microbiome, with predominating Pseudomonas, followed by the Spiroplasma, Apibacter, Enterococcus, Dysgonomonas, Sebaldella, Ruminococcus, and Wolbachia. Interesting dissimilarities were also discovered within and between the beetle microbiomes in terms of genders and organs. Analyses showed that Spiroplasma / Apibacter as well as Pseudomonas / Pseudomonas were the most abundant in the genitals / intestines of male and female beetles, respectively. Bacterial richness did not display any significant difference in the three provinces but was higher in male beetles than in females and more in the genitals than intestines.
Conclusions: The present study identified Pseudomonas-like endobacterium as a common symbiont of P. fuscipes beetles; this bacterium begins its journey from gut and genitalia of females to reach the male rove beetles. Additionally, male and female rove beetles were characterized by distinctive microbiota in different organs, likely reflecting different functions and/or adaptation processes. Evidence of the extension of P. fuscipes microbiome from the environmental paradigm to the pathobiome was also presented herein. A comprehensive survey of P. fuscipes microbiome components may eventually lead to ecological insights into the production and utilization of defensive compound of pederin and also the management of linear dermatitis with the use of available antibiotics against bacterial pathogens released by the beetles. Video Abstract.
Keywords: Apibacter; Dermatitis linearis; Genital microbiota; Gut microbiota; Pederin; Pseudomonas-like Paederus fuscipes endosymbiont; Spiroplasma; Wolbachia.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- van Huis A. The global impact of insects. Wageningen University, Wageningen UR; 2014.
-
- Maleki-Ravasan N, Oshaghi MA, Afshar D, Arandian MH, Hajikhani S, Akhavan AA, Yakhchali B, Shirazi MH, Rassi Y, Jafari R, et al. Aerobic bacterial flora of biotic and abiotic compartments of a hyperendemic Zoonotic Cutaneous Leishmaniasis (ZCL) focus. Parasites Vectors. 2015;8:1–22. doi: 10.1186/s13071-014-0517-3. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

