Phytochrome F mediates red light responsiveness additively with phytochromes B1 and B2 in tomato
- PMID: 36670526
- PMCID: PMC10069882
- DOI: 10.1093/plphys/kiad028
Phytochrome F mediates red light responsiveness additively with phytochromes B1 and B2 in tomato
Abstract
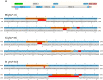
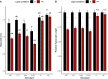
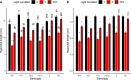
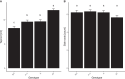
Phytochromes are red light and far-red light sensitive, plant-specific light receptors that allow plants to orient themselves in space and time. Tomato (Solanum lycopersicum) contains a small family of five phytochrome genes, for which to date stable knockout mutants are only available for three of them. Using CRISPR technology, we created multiple alleles of SlPHYTOCHROME F (phyF) mutants to determine the function of this understudied phytochrome. We report that SlphyF acts as a red/far-red light reversible low fluence sensor, likely through the formation of heterodimers with SlphyB1 and SlphyB2. During photomorphogenesis, phyF functions additively with phyB1 and phyB2. Our data further suggest that phyB2 requires the presence of either phyB1 or phyF during seedling de-etiolation in red light, probably via heterodimerization, while phyB1 homodimers are required and sufficient to suppress hypocotyl elongation in red light. During the end-of-day far-red response, phyF works additively with phyB1 and phyB2. In addition, phyF plays a redundant role with phyB1 in photoperiod detection and acts additively with phyA in root patterning. Taken together, our results demonstrate various roles for SlphyF during seedling establishment, sometimes acting additively, other times acting redundantly with the other phytochromes in tomato.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




References
-
- Alba R, Kelmenson PM, Cordonnier-Pratt M-M, Pratt LH (2000) The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol Biol Evol 17(3): 362–373 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

