A Two-Compartment Fermentation System to Quantify Strain-Specific Interactions in Microbial Co-Cultures
- PMID: 36671675
- PMCID: PMC9854596
- DOI: 10.3390/bioengineering10010103
A Two-Compartment Fermentation System to Quantify Strain-Specific Interactions in Microbial Co-Cultures
Abstract
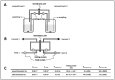
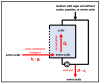
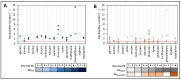
To fulfil the growing interest in investigating microbial interactions in co-cultures, a novel two-compartment bioreactor system was developed, characterised, and implemented. The system allowed for the exchange of amino acids and peptides via a polyethersulfone membrane that retained biomass. Further system characterisation revealed a Bodenstein number of 18, which hints at backmixing. Together with other physical settings, the existence of unwanted inner-compartment substrate gradients could be ruled out. Furthermore, the study of Damkoehler numbers indicated that a proper metabolite supply between compartments was enabled. Implementing the two-compartment system (2cs) for growing Streptococcus thermophilus and Lactobacillus delbrueckii subs. bulgaricus, which are microorganisms commonly used in yogurt starter cultures, revealed only a small variance between the one-compartment and two-compartment approaches. The 2cs enabled the quantification of the strain-specific production and consumption rates of amino acids in an interacting S. thermophilus-L. bulgaricus co-culture. Therefore, comparisons between mono- and co-culture performance could be achieved. Both species produce and release amino acids. Only alanine was produced de novo from glucose through potential transaminase activity by L. bulgaricus and consumed by S. thermophilus. Arginine availability in peptides was limited to S. thermophilus' growth, indicating active biosynthesis and dependency on the proteolytic activity of L. bulgaricus. The application of the 2cs not only opens the door for the quantification of exchange fluxes between microbes but also enables continuous production modes, for example, for targeted evolution studies.
Keywords: Lactobacillus bulgaricus; Streptococcus thermophilus; bioprocess engineering; lactic acid bacteria; metabolomics; microbial consortia.
Conflict of interest statement
The authors declare no financial or commercial conflicts of interest.
Figures





References
-
- West S.A., Diggle S.P., Buckling A., Gardner A., Griffin A.S. The Social Lives of Microbes. Annu. Rev. Ecol. Evol. Syst. 2007;38:53–77. doi: 10.1146/annurev.ecolsys.38.091206.095740. - DOI
-
- Ziesack M., Gibson T., Oliver J.K.W., Shumaker A.M., Hsu B.B., Riglar D.T., Giessen T.W., Di Benedetto N.V., Bry L., Way J.C., et al. Engineered Interspecies Amino Acid Cross-Feeding Increases Population Evenness in a Synthetic Bacterial Consortium. mSystems. 2019;4:e00352-19. doi: 10.1128/mSystems.00352-19. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

