Optimization of 3D Immunofluorescence Analysis and Visualization Using IMARIS and MeshLab
- PMID: 36672153
- PMCID: PMC9856541
- DOI: 10.3390/cells12020218
Optimization of 3D Immunofluorescence Analysis and Visualization Using IMARIS and MeshLab
Abstract
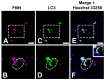
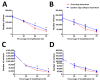
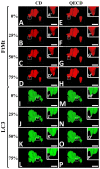
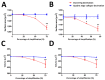
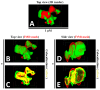
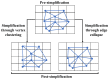
The precision of colocalization analysis is enhanced by 3D and is potentially more accurate than 2D. Even though 3D improves the visualization of colocalization analysis, rendering a colocalization model may generate a model with numerous polygons. We developed a 3D colocalization model of FtMt/LC3 followed by simplification. Double immunofluorescence staining of FtMt and LC3 was conducted, and stacked images were acquired. We used IMARIS to render the 3D colocalization model of FtMt/LC3 and further processed it with MeshLab to decimate and generate a less complex colocalization model. We examined the available simplification algorithm using MeshLab in detail and evaluated the feasibility of each procedure in generating a model with less complexity. The quality of the simplified model was subsequently assessed. MeshLab's available shaders were scrutinized to facilitate the spatial colocalization determination. Finally, we showed that QECD was the most effective method for reducing the polygonal complexity of the colocalization model without compromising its quality. In addition, we would recommend implementing the x-ray shader, which we found useful for visualizing colocalization. As 3D was found to be more accurate in quantifying colocalization, our study provides a novel and dependable method for rendering 3D models for colocalization analysis.
Keywords: 3D; IMARIS; MeshLab; colocalization analysis; decimation; simplification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Abu Bakar Z.H., Bellier J.-P., Yanagisawa D., Kato T., Mukaisho K.-i., Tooyama I. LC3/FtMt colocalization patterns reveal the progression of FtMt accumulation in nigral neurons of patients with progressive supranuclear palsy. Int. J. Mol. Sci. 2022;23:537. doi: 10.3390/ijms23010537. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

