Genome-Wide Methylation Changes Associated with Replicative Senescence and Differentiation in Endothelial and Bone Marrow Mesenchymal Stromal Cells
- PMID: 36672222
- PMCID: PMC9857206
- DOI: 10.3390/cells12020285
Genome-Wide Methylation Changes Associated with Replicative Senescence and Differentiation in Endothelial and Bone Marrow Mesenchymal Stromal Cells
Abstract
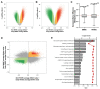
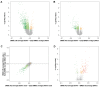
Bone marrow mesenchymal stromal cells (BMSCs) are multipotent cells able to self-renew and differentiate, depending on the microenvironment, into adipocytes and osteoblasts. These cells have a limited number of replications and enter replicative senescence during in vitro expansion. The role of DNA methylation (DNAm) assumes importance in cell function and commitment; however, its exact contribution to BMSC differentiation and replicative senescence is still unclear. We performed a genome-wide DNAm analysis on BMSCs cultured in vitro at early passages and induced to differentiate into adipocytes and osteoblasts, and on replicative senescent BMSCs and HUVECs, to identify DNAm patterns of senescence and differentiation. We also compared BMSCs and HUVECs in replicative senescence and found that, in both cellular systems, genome-wide hypomethylation was accompanied by a higher-than-expected overlap of differentially methylated positions (DMPs) and concordance in terms of direction of the change. A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on lineage-independent senescence-associated DMPs revealed 16 common pathways, including Insulin resistance, Molecule adhesion, and Wnt/β-catenin signaling. In both adipogenesis and osteogenesis, we observed a general demethylation of CpG sites compared with undifferentiated BMSCs with a higher number of DMPs in osteogenesis. KEGG analysis resulted in 30 pathways enriched in osteoblasts and only 2 in adipocytes when compared to undifferentiated cells. When comparing differentiated BMSCs with senescent ones, osteogenesis exhibited a greater overlap with senescence in terms of number of DMPs and direction of methylation change compared to adipogenesis. In conclusion, this study may be useful for future research on general mechanisms that occur in replicative senescence and furthermore to identify trajectories of BMSC differentiation and common aspects of differentiated and senescent cells.
Keywords: DNA methylation; adipocytes; bone-marrow mesenchymal stromal cells; cellular senescence; endothelial cells; osteoblasts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

