Multiomics Study of a Novel Naturally Derived Small Molecule, NSC772864, as a Potential Inhibitor of Proto-Oncogenes Regulating Cell Cycle Progression in Colorectal Cancer
- PMID: 36672275
- PMCID: PMC9856482
- DOI: 10.3390/cells12020340
Multiomics Study of a Novel Naturally Derived Small Molecule, NSC772864, as a Potential Inhibitor of Proto-Oncogenes Regulating Cell Cycle Progression in Colorectal Cancer
Abstract
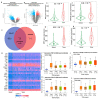
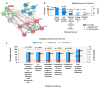
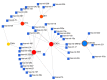
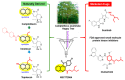
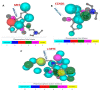
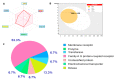
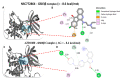
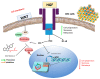
Colorectal cancer (CRC) is one of the most prevalent malignant tumors, and it contributes to high numbers of deaths globally. Although advances in understanding CRC molecular mechanisms have shed significant light on its pathogenicity, current treatment options, including combined chemotherapy and molecular-targeted agents, are still limited due to resistance, with almost 25% of patients developing distant metastasis. Therefore, identifying novel biomarkers for early diagnosis is crucial, as they will also influence strategies for new targeted therapies. The proto-oncogene, c-Met, a tyrosine kinase that promotes cell proliferation, motility, and invasion; c-MYC, a transcription factor associated with the modulation of the cell cycle, proliferation, apoptosis; and cyclin D1 (CCND1), an essential regulatory protein in the cell cycle, all play crucial roles in cancer progression. In the present study, we explored computational simulations through bioinformatics analysis and identified the overexpression of c-Met/GSK3β/MYC/CCND1 oncogenic signatures that were associated with cancer progression, drug resistance, metastasis, and poor clinical outcomes in CRC. We further demonstrated the anticancer activities of our newly synthesized quinoline-derived compound, NSC772864, against panels of the National Cancer Institute's human CRC cell lines. The compound exhibited cytotoxic activities against various CRC cell lines. Using target prediction tools, we found that c-Met/GSK3β/MYC/CCND1 were target genes for the NSC772864 compound. Subsequently, we performed in silico molecular docking to investigate protein-ligand interactions and discovered that NSC772864 exhibited higher binding affinities with these oncogenes compared to FDA-approved drugs. These findings strongly suggest that NSC772864 is a novel and potential antiCRC agent.
Keywords: colorectal cancer; drug resistance; molecular docking simulation; protein–ligand interaction; small molecule.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Comprehensive Omics Analysis of a Novel Small-Molecule Inhibitor of Chemoresistant Oncogenic Signatures in Colorectal Cancer Cell with Antitumor Effects.Cells. 2021 Aug 3;10(8):1970. doi: 10.3390/cells10081970. Cells. 2021. PMID: 34440739 Free PMC article.
-
Novel oncogene 5MP1 reprograms c-Myc translation initiation to drive malignant phenotypes in colorectal cancer.EBioMedicine. 2019 Jun;44:387-402. doi: 10.1016/j.ebiom.2019.05.058. Epub 2019 Jun 4. EBioMedicine. 2019. PMID: 31175057 Free PMC article.
-
Disruption of the c-Myc/miR-200b-3p/PRDX2 regulatory loop enhances tumor metastasis and chemotherapeutic resistance in colorectal cancer.J Transl Med. 2017 Dec 19;15(1):257. doi: 10.1186/s12967-017-1357-7. J Transl Med. 2017. PMID: 29258530 Free PMC article.
-
Small-molecule drugs of colorectal cancer: Current status and future directions.Biochim Biophys Acta Mol Basis Dis. 2024 Jan;1870(1):166880. doi: 10.1016/j.bbadis.2023.166880. Epub 2023 Sep 9. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 37696461 Review.
-
Copper in colorectal cancer: From copper-related mechanisms to clinical cancer therapies.Clin Transl Med. 2024 Jun;14(6):e1724. doi: 10.1002/ctm2.1724. Clin Transl Med. 2024. PMID: 38804588 Free PMC article. Review.
Cited by
-
Rewired Metabolism Caused by the Oncogenic Deregulation of MYC as an Attractive Therapeutic Target in Cancers.Cells. 2023 Jun 29;12(13):1745. doi: 10.3390/cells12131745. Cells. 2023. PMID: 37443779 Free PMC article. Review.
-
Comprehensive analysis of bulk and single-cell RNA sequencing data reveals Schlafen-5 (SLFN5) as a novel prognosis and immunity.Int J Med Sci. 2024 Sep 9;21(12):2348-2364. doi: 10.7150/ijms.97975. eCollection 2024. Int J Med Sci. 2024. PMID: 39310264 Free PMC article.
-
Uncovering the anti-cancer mechanism of cucurbitacin D against colorectal cancer through network pharmacology and molecular docking.Discov Oncol. 2025 Apr 17;16(1):551. doi: 10.1007/s12672-025-02056-7. Discov Oncol. 2025. PMID: 40244518 Free PMC article.
-
Spermine Enhances the Peroxidase Activities of Multimeric Antiparallel G-quadruplex DNAzymes.Biosensors (Basel). 2025 Jan 2;15(1):12. doi: 10.3390/bios15010012. Biosensors (Basel). 2025. PMID: 39852063 Free PMC article.
References
-
- Van Cutsem E., Köhne C.H., Láng I., Folprecht G., Nowacki M.P., Cascinu S., Shchepotin I., Maurel J., Cunningham D., Tejpar S., et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: Updated analysis of overall survival according to tumor K.R.A.S. and B.R.A.F. mutation status. J. Clin. Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

