Serum Mass Spectrometry Proteomics and Protein Set Identification in Response to FOLFOX-4 in Drug-Resistant Ovarian Carcinoma
- PMID: 36672361
- PMCID: PMC9856519
- DOI: 10.3390/cancers15020412
Serum Mass Spectrometry Proteomics and Protein Set Identification in Response to FOLFOX-4 in Drug-Resistant Ovarian Carcinoma
Abstract
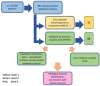
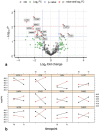
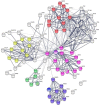
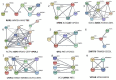
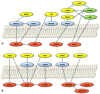
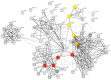
Ovarian cancer is a highly lethal gynecological malignancy. Drug resistance rapidly occurs, and different therapeutic approaches are needed. So far, no biomarkers have been discovered to predict early response to therapies in the case of multi-treated ovarian cancer patients. The aim of our investigation was to identify a protein panel and the molecular pathways involved in chemotherapy response through a combination of studying proteomics and network enrichment analysis by considering a subset of samples from a clinical setting. Differential mass spectrometry studies were performed on 14 serum samples from patients with heavily pretreated platinum-resistant ovarian cancer who received the FOLFOX-4 regimen as a salvage therapy. The serum was analyzed at baseline time (T0) before FOLFOX-4 treatment, and before the second cycle of treatment (T1), with the aim of understanding if it was possible, after a first treatment cycle, to detect significant proteome changes that could be associated with patients responses to therapy. A total of 291 shared expressed proteins was identified and 12 proteins were finally selected between patients who attained partial response or no-response to chemotherapy when both response to therapy and time dependence (T0, T1) were considered in the statistical analysis. The protein panel included APOL1, GSN, GFI1, LCATL, MNA, LYVE1, ROR1, SHBG, SOD3, TEC, VPS18, and ZNF573. Using a bioinformatics network enrichment approach and metanalysis study, relationships between serum and cellular proteins were identified. An analysis of protein networks was conducted and identified at least three biological processes with functional and therapeutic significance in ovarian cancer, including lipoproteins metabolic process, structural component modulation in relation to cellular apoptosis and autophagy, and cellular oxidative stress response. Five proteins were almost independent from the network (LYVE1, ROR1, TEC, GFI1, and ZNF573). All proteins were associated with response to drug-resistant ovarian cancer resistant and were mechanistically connected to the pathways associated with cancer arrest. These results can be the basis for extending a biomarker discovery process to a clinical trial, as an early predictive tool of chemo-response to FOLFOX-4 of heavily treated ovarian cancer patients and for supporting the oncologist to continue or to interrupt the therapy.
Keywords: FOLFOX-4; cancer molecular pathways; mass spectrometry proteomics; network enrichment analysis; ovarian cancer; protein panel; serum samples; time lapse detection.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Raymond E., Buquet-Fagot C., Djelloul S., Mester J., Cvitkovic E., Allain P., Louvet C., Gespach C. Antitumor Activity of Oxaliplatin in Combination with 5-Fluorouracil and the Thymidylate Synthase Inhibitor AG337 in Human Colon, Breast and Ovarian Cancers. Anticancer. Drugs. 1997;8:876–885. doi: 10.1097/00001813-199710000-00009. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

