Approach to Cohort-Wide Re-Analysis of Exome Data in 1000 Individuals with Neurodevelopmental Disorders
- PMID: 36672771
- PMCID: PMC9858523
- DOI: 10.3390/genes14010030
Approach to Cohort-Wide Re-Analysis of Exome Data in 1000 Individuals with Neurodevelopmental Disorders
Abstract
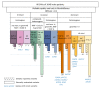
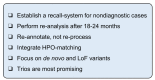
The re-analysis of nondiagnostic exome sequencing (ES) has the potential to increase diagnostic yields in individuals with rare diseases, but its implementation in the daily routines of laboratories is limited due to restricted capacities. Here, we describe a systematic approach to re-analyse the ES data of a cohort consisting of 1040 diagnostic and nondiagnostic samples. We applied a strict filter cascade to reveal the most promising single-nucleotide variants (SNVs) of the whole cohort, which led to an average of 0.77 variants per individual that had to be manually evaluated. This variant set revealed seven novel diagnoses (0.8% of all nondiagnostic cases) and two secondary findings. Thirteen additional variants were identified by a scientific approach prior to this re-analysis and were also present in this variant set. This resulted in a total increase in the diagnostic yield of 2.3%. The filter cascade was optimised during the course of the study and finally resulted in sensitivity of 85%. After applying the filter cascade, our re-analysis took 20 h and enabled a workflow that can be used repeatedly. This work is intended to provide a practical recommendation for other laboratories wishing to introduce a resource-efficient re-analysis strategy into their clinical routine.
Keywords: exome sequencing; neurodevelopmental disorder; re-analysis.
Conflict of interest statement
T.H. is employed by Limbus, Medical Technologies GmbH, Rostock, Germany. The other authors declare no conflict of interest.
Figures


References
-
- Deignan J.L., Chung W.K., Kearney H.M., Monaghan K.G., Rehder C.W., Chao E.C. Points to Consider in the Reevaluation and Reanalysis of Genomic Test Results: A Statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2019;21:1267–1270. doi: 10.1038/s41436-019-0478-1. - DOI - PMC - PubMed
-
- Dai P., Honda A., Ewans L., McGaughran J., Burnett L., Law M., Phan T.G. Recommendations for next Generation Sequencing Data Reanalysis of Unsolved Cases with Suspected Mendelian Disorders: A Systematic Review and Meta-Analysis. Genet. Med. 2022;24:1618–1629. doi: 10.1016/j.gim.2022.04.021. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

