Characteristics and Expression Analysis of Invertase Gene Family in Common Wheat (Triticum aestivum L.)
- PMID: 36672783
- PMCID: PMC9858860
- DOI: 10.3390/genes14010041
Characteristics and Expression Analysis of Invertase Gene Family in Common Wheat (Triticum aestivum L.)
Abstract
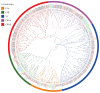
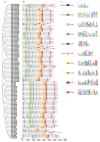
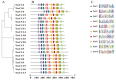
Invertase (INV) irreversibly catalyzes the conversion of sucrose into glucose and fructose, playing important role in plant development and stress tolerance. However, the functions of INV genes in wheat have been less studied. In this study, a total of 126 TaINV genes were identified using a genome-wide search method, which could be classified into five classes (TaCWI-α, TaCWI-β, TaCI-α, TaCI-β, and TaVI) based on phylogenetic relationship. A total of 101 TaINVs were collinear with their ancestors in the synteny analysis, and we speculated that polyploidy events were the main force in the expansion of the TaINV gene family. Compared with TaCI, TaCWI and TaVI are more similar in gene structure and protein properties. Transcriptome sequencing analysis showed that TaINVs expressed in multiple tissues with different expression levels. Among 19 tissue-specific expressed TaINVs, 12 TaINVs showed grain-specific expression pattern and might play an important role in wheat grain development. In addition, qRT-PCR results further confirmed that TaCWI50 and TaVI27 show different expression in grain weight NILs. Our results demonstrated that the high expression of TaCWI50 and TaVI27 may be associated with a larger TGW phenotype. This work provides the foundations for understanding the grain development mechanism.
Keywords: expression profiles; genome-wide; invertase; wheat.
Conflict of interest statement
Informed consent was obtained from all subjects involved in the study.
Figures






Similar articles
-
A yield-associated gene TaCWI, in wheat: its function, selection and evolution in global breeding revealed by haplotype analysis.Theor Appl Genet. 2015 Jan;128(1):131-43. doi: 10.1007/s00122-014-2417-5. Epub 2014 Nov 4. Theor Appl Genet. 2015. PMID: 25367379
-
Genome-Wide Identification and Expression Profiles of Late Embryogenesis-Abundant (LEA) Genes during Grain Maturation in Wheat (Triticum aestivum L.).Genes (Basel). 2019 Sep 10;10(9):696. doi: 10.3390/genes10090696. Genes (Basel). 2019. PMID: 31510067 Free PMC article.
-
Comprehensive analysis of polygalacturonase gene family highlights candidate genes related to pollen development and male fertility in wheat (Triticum aestivum L.).Planta. 2020 Aug 1;252(2):31. doi: 10.1007/s00425-020-03435-w. Planta. 2020. PMID: 32740680
-
TaFlo2-A1, an ortholog of rice Flo2, is associated with thousand grain weight in bread wheat (Triticum aestivum L.).BMC Plant Biol. 2017 Oct 16;17(1):164. doi: 10.1186/s12870-017-1114-3. BMC Plant Biol. 2017. PMID: 29037166 Free PMC article.
-
Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.).BMC Genomics. 2019 Feb 1;20(1):101. doi: 10.1186/s12864-019-5455-1. BMC Genomics. 2019. PMID: 30709338 Free PMC article.
Cited by
-
Grain under pressure: Harnessing biochemical pathways to beat drought and heat in wheat.Plant J. 2025 Jun;122(6):e70253. doi: 10.1111/tpj.70253. Plant J. 2025. PMID: 40532117 Free PMC article. Review.
-
Genome-Wide Comparative Analysis of Invertases in the Salicaceae with the Identification of Genes Involved in Catkin Fiber Initiation and Development.Curr Issues Mol Biol. 2025 Jun 5;47(6):423. doi: 10.3390/cimb47060423. Curr Issues Mol Biol. 2025. PMID: 40699822 Free PMC article.
-
Genome-wide characterization of FK506-binding proteins, parvulins and phospho-tyrosyl phosphatase activators in wheat and their regulation by heat stress.Front Plant Sci. 2022 Dec 15;13:1053524. doi: 10.3389/fpls.2022.1053524. eCollection 2022. Front Plant Sci. 2022. PMID: 36589073 Free PMC article.
-
Exploring the role of GhN/AINV23: implications for plant growth, development, and drought tolerance.Biol Direct. 2024 Mar 14;19(1):22. doi: 10.1186/s13062-024-00465-2. Biol Direct. 2024. PMID: 38486336 Free PMC article.
-
N-acetylcysteine mitigates oxidative damage to the ovary in D-galactose-induced ovarian failure in rabbits.Mol Biol Rep. 2024 Sep 23;51(1):1008. doi: 10.1007/s11033-024-09951-2. Mol Biol Rep. 2024. PMID: 39312076
References
-
- Haouazine-Takvorian N., Tymowska-Lalanne Z., Takvorian A., Tregear J., Lejeune B., Alain Lecharny A., Kreis M. Characterization of two members of the Arabidopsis thaliana gene family, Atβfruct3 and Atβfruct4, coding for vacuolar invertases. Gene. 1997;197:239–251. doi: 10.1016/S0378-1119(97)00268-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

