Genomic Regions Associated with Wool, Growth and Reproduction Traits in Uruguayan Merino Sheep
- PMID: 36672908
- PMCID: PMC9858812
- DOI: 10.3390/genes14010167
Genomic Regions Associated with Wool, Growth and Reproduction Traits in Uruguayan Merino Sheep
Abstract
The aim of this study was to identify genomic regions and genes associated with the fiber diameter (FD), clean fleece weight (CFW), live weight (LW), body condition score (BCS), pregnancy rate (PR) and lambing potential (LP) of Uruguayan Merino sheep. Phenotypic records of approximately 2000 mixed-age ewes were obtained from a Merino nucleus flock. Genome-wide association studies were performed utilizing single-step Bayesian analysis. For wool traits, a total of 35 genomic windows surpassed the significance threshold (PVE ≥ 0.25%). The proportion of the total additive genetic variance explained by those windows was 4.85 and 9.06% for FD and CFW, respectively. There were 42 windows significantly associated with LWM, which collectively explained 43.2% of the additive genetic variance. For BCS, 22 relevant windows accounted for more than 40% of the additive genetic variance, whereas for the reproduction traits, 53 genomic windows (24 and 29 for PR and LP, respectively) reached the suggestive threshold of 0.25% of the PVE. Within the top 10 windows for each trait, we identified several genes showing potential associations with the wool (e.g., IGF-1, TGFB2R, PRKCA), live weight (e.g., CAST, LAP3, MED28, HERC6), body condition score (e.g., CDH10, TMC2, SIRPA, CPXM1) or reproduction traits (e.g., ADCY1, LEPR, GHR, LPAR2) of the mixed-age ewes.
Keywords: GWAS; body condition score; fiber diameter; gene; reproduction; sheep.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
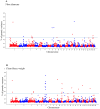

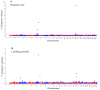
References
-
- Fernando R., Toosi A., Wolc A., Garrick D., Dekkers J. Application of whole-genome prediction methods for genome-wide association studies: A Bayesian approach. J. Agric. Biol. Environ. Stat. 2017;22:172–193. doi: 10.1007/s13253-017-0277-6. - DOI
-
- Fernando R.L., Garrick D. Genome-Wide Association Studies and Genomic Prediction. Humana Press; Totowa, NJ, USA: 2013. Bayesian methods applied to GWAS; pp. 237–274. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

