Development and Assessment of a Novel Core Biopsy-Based Prediction Model for Pathological Complete Response to Neoadjuvant Chemotherapy in Women with Breast Cancer
- PMID: 36674372
- PMCID: PMC9867383
- DOI: 10.3390/ijerph20021617
Development and Assessment of a Novel Core Biopsy-Based Prediction Model for Pathological Complete Response to Neoadjuvant Chemotherapy in Women with Breast Cancer
Abstract
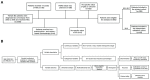
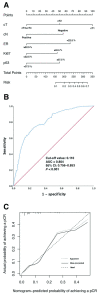
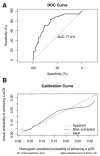
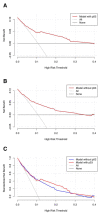
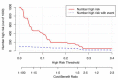
Purpose: Pathological complete response (pCR), the goal of NAC, is considered a surrogate for favorable outcomes in breast cancer (BC) patients administrated neoadjuvant chemotherapy (NAC). This study aimed to develop and assess a novel nomogram model for predicting the probability of pCR based on the core biopsy. Methods: This was a retrospective study involving 920 BC patients administered NAC between January 2012 and December 2018. The patients were divided into a primary cohort (769 patients from January 2012 to December 2017) and a validation cohort (151 patients from January 2017 to December 2018). After converting continuous variables to categorical variables, variables entering the model were sequentially identified via univariate analysis, a multicollinearity test, and binary logistic regression analysis, and then, a nomogram model was developed. The performance of the model was assessed concerning its discrimination, accuracy, and clinical utility. Results: The optimal predictive threshold for estrogen receptor (ER), Ki67, and p53 were 22.5%, 32.5%, and 37.5%, respectively (all p < 0.001). Five variables were selected to develop the model: clinical T staging (cT), clinical nodal (cN) status, ER status, Ki67 status, and p53 status (all p ≤ 0.001). The nomogram showed good discrimination with the area under the curve (AUC) of 0.804 and 0.774 for the primary and validation cohorts, respectively, and good calibration. Decision curve analysis (DCA) showed that the model had practical clinical value. Conclusions: This study constructed a novel nomogram model based on cT, cN, ER status, Ki67 status, and p53 status, which could be applied to personalize the prediction of pCR in BC patients treated with NAC.
Keywords: breast cancer; neoadjuvant chemotherapy; nomogram; pathological complete response; prediction model.
Conflict of interest statement
The authors declare that the research was conducted with no commercial or financial relationships that could be construed as potential conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

