Identification of the Actin-Binding Region and Binding to Host Plant Apple Actin of Immunodominant Transmembrane Protein of ' Candidatus Phytoplasma mali'
- PMID: 36674483
- PMCID: PMC9860668
- DOI: 10.3390/ijms24020968
Identification of the Actin-Binding Region and Binding to Host Plant Apple Actin of Immunodominant Transmembrane Protein of ' Candidatus Phytoplasma mali'
Abstract
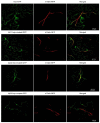
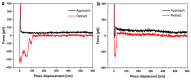
'Candidatus Phytoplasma mali' ('Ca. P. mali') has only one major membrane protein, the immunodominant membrane protein (Imp), which is regarded as being close to the ancestor of all phytoplasma immunodominant membrane proteins. Imp binds to actin and possibly facilitates its movement in the plant or insect host cells. However, protein sequences of Imp are quite diverse among phytoplasma species, thus resulting in difficulties in identifying conserved domains across species. In this work, we compare Imp protein sequences of 'Ca. P. mali' strain PM19 (Imp-PM19) with Imp of different strains of 'Ca. P. mali' and identify its actin-binding domain. Moreover, we show that Imp binds to the actin of apple (Malus x domestica), which is the host plant of 'Ca. P. mali'. Using molecular and scanning force spectroscopy analysis, we find that the actin-binding domain of Imp-PM19 contains a highly positively charged amino acid cluster. Our result could allow investigating a possible correlation between Imp variants and the infectivity of the corresponding 'Ca. P. mali' isolates.
Keywords: actin; immunodominant membrane protein; phytoplasma; single-molecule force spectroscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Ding L., Lu W., Yang Y., Zhong Q., Zhou T., Wang G., Song C., Ma W., Chen W., Wu Y. Immunodominant membrane protein (Imp) promotes the transmission of wheat blue dwarf (WBD) phytoplasma by directly interacting with α-tubulin in leafhoppers. Eur. J. Plant Pathol. 2022;162:357–367. doi: 10.1007/s10658-021-02408-3. - DOI
-
- Strohmayer A., Schwarz T., Braun M., Krczal G., Boonrod K. The Effect of the Anticipated Nuclear Localization Sequence of ‘Candidatus Phytoplasma mali’ SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor. Microorganisms. 2021;9:1756. doi: 10.3390/microorganisms9081756. - DOI - PMC - PubMed
-
- Clark M.F., Morton A., Buss S.L. Preparation of mycoplasma immunogens from plants and a comparison of polyclonal and monoclonal antibodies made against primula yellows MLO-associated antigens. Ann. Appl. Biol. 1989;114:111–124. doi: 10.1111/j.1744-7348.1989.tb06791.x. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

