CRISPR/Cas9-Mediated Enrichment Coupled to Nanopore Sequencing Provides a Valuable Tool for the Precise Reconstruction of Large Genomic Target Regions
- PMID: 36674592
- PMCID: PMC9863143
- DOI: 10.3390/ijms24021076
CRISPR/Cas9-Mediated Enrichment Coupled to Nanopore Sequencing Provides a Valuable Tool for the Precise Reconstruction of Large Genomic Target Regions
Abstract
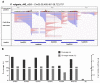
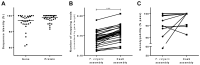
Complete and accurate identification of genetic variants associated with specific phenotypes can be challenging when there is a high level of genomic divergence between individuals in a study and the corresponding reference genome. We have applied the Cas9-mediated enrichment coupled to nanopore sequencing to perform a targeted de novo assembly and accurately reconstruct a genomic region of interest. This approach was used to reconstruct a 250-kbp target region on chromosome 5 of the common bean genome (Phaseolus vulgaris) associated with the shattering phenotype. Comparing a non-shattering cultivar (Midas) with the reference genome revealed many single-nucleotide variants and structural variants in this region. We cut five 50-kbp tiled sub-regions of Midas genomic DNA using Cas9, followed by sequencing on a MinION device and de novo assembly, generating a single contig spanning the whole 250-kbp region. This assembly increased the number of Illumina reads mapping to genes in the region, improving their genotypability for downstream analysis. The Cas9 tiling approach for target enrichment and sequencing is a valuable alternative to whole-genome sequencing for the assembly of ultra-long regions of interest, improving the accuracy of downstream genotype-phenotype association analysis.
Keywords: Cas9-tiling enrichment; de novo assembly; nanopore sequencing; pod-shattering; variant calling.
Conflict of interest statement
Authors M.R. and M.D. are partners of Genartis srl. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Maestri S., Gambino G., Lopatriello G., Minio A., Perrone I., Cosentino E., Giovannone B., Marcolungo L., Alfano M., Rombauts S., et al. ‘Nebbiolo’ Genome Assembly Allows Surveying the Occurrence and Functional Implications of Genomic Structural Variations in Grapevines (Vitis vinifera L.) BMC Genom. 2022;23:159. doi: 10.1186/s12864-022-08389-9. - DOI - PMC - PubMed
-
- Valiente-Mullor C., Beamud B., Ansari I., Francés-Cuesta C., García-González N., Mejía L., Ruiz-Hueso P., González-Candelas F. One Is Not Enough: On the Effects of Reference Genome for the Mapping and Subsequent Analyses of Short-Reads. PLoS Comput. Biol. 2021;17:e1008678. doi: 10.1371/journal.pcbi.1008678. - DOI - PMC - PubMed

