Peculiar Ca2+ Homeostasis, ER Stress, Autophagy, and TG2 Modulation in Celiac Disease Patient-Derived Cells
- PMID: 36675008
- PMCID: PMC9866799
- DOI: 10.3390/ijms24021495
Peculiar Ca2+ Homeostasis, ER Stress, Autophagy, and TG2 Modulation in Celiac Disease Patient-Derived Cells
Abstract
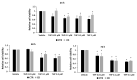
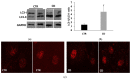
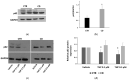
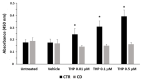
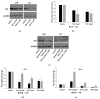
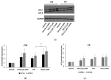
Celiac disease (CD) is an inflammatory intestinal disease caused by the ingestion of gluten-containing cereals by genetically predisposed individuals. Constitutive differences between cells from CD patients and control subjects, including levels of protein phosphorylation, alterations of vesicular trafficking, and regulation of type 2 transglutaminase (TG2), have been reported. In the present work, we investigated how skin-derived fibroblasts from CD and control subjects responded to thapsigargin, an endoplasmic reticulum ER stress inducer, in an attempt to contribute to the comprehension of molecular features of the CD cellular phenotype. We analyzed Ca2+ levels by single-cell video-imaging and TG2 activity by a microplate assay. Western blots and PCR analyses were employed to monitor TG2 levels and markers of ER stress and autophagy. We found that the cytosolic and ER Ca2+ level of CD cells was lower than in control cells. Treatments with thapsigargin differently activated TG2 in control and CD cells, as well as caused slightly different responses regarding the activation of ER stress and the expression of autophagic markers. On the whole, our findings identified further molecular features of the celiac cellular phenotype and highlighted that CD cells appeared less capable of adapting to a stress condition and responding in a physiological way.
Keywords: Ca2+ homeostasis; ER stress; autophagy; celiac disease; thapsigargin; type 2 transglutaminase; unfolded protein response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



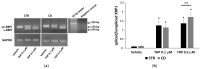





Similar articles
-
Gliadin-dependent UPR induction directly triggers the expression of TG2 and pro-inflammatory cytokines, dysregulates intestinal permeability, and reduces CFTR expression in intestinal epithelial cells of celiac disease patients.Biol Direct. 2025 Apr 17;20(1):55. doi: 10.1186/s13062-025-00644-9. Biol Direct. 2025. PMID: 40247380 Free PMC article.
-
Constitutive Differential Features of Type 2 Transglutaminase in Cells Derived from Celiac Patients and from Healthy Subjects.Int J Mol Sci. 2020 Feb 12;21(4):1231. doi: 10.3390/ijms21041231. Int J Mol Sci. 2020. PMID: 32059410 Free PMC article.
-
Celiac anti-type 2 transglutaminase antibodies induce differential effects in fibroblasts from celiac disease patients and from healthy subjects.Amino Acids. 2017 Mar;49(3):541-550. doi: 10.1007/s00726-016-2307-z. Epub 2016 Sep 9. Amino Acids. 2017. PMID: 27613408
-
Type 2 Transglutaminase in Coeliac Disease: A Key Player in Pathogenesis, Diagnosis and Therapy.Int J Mol Sci. 2022 Jul 6;23(14):7513. doi: 10.3390/ijms23147513. Int J Mol Sci. 2022. PMID: 35886862 Free PMC article. Review.
-
Transglutaminase 2 and Transglutaminase 2 Autoantibodies in Celiac Disease: a Review.Clin Rev Allergy Immunol. 2019 Aug;57(1):23-38. doi: 10.1007/s12016-016-8557-4. Clin Rev Allergy Immunol. 2019. PMID: 27263022 Review.
Cited by
-
3,3'-Diindolylmethane inhibits the proliferation of esophageal squamous cell carcinoma cells via downregulation of STIM1.Oncol Lett. 2024 May 28;28(2):339. doi: 10.3892/ol.2024.14473. eCollection 2024 Aug. Oncol Lett. 2024. PMID: 38855503 Free PMC article.
-
Psychiatric and Neurological Manifestations of Celiac Disease in Adults.Cureus. 2023 Mar 3;15(3):e35712. doi: 10.7759/cureus.35712. eCollection 2023 Mar. Cureus. 2023. PMID: 36875248 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

