Reprogramming of Fundamental miRNA and Gene Expression during the Barley- Piriformospora indica Interaction
- PMID: 36675845
- PMCID: PMC9865155
- DOI: 10.3390/jof9010024
Reprogramming of Fundamental miRNA and Gene Expression during the Barley- Piriformospora indica Interaction
Abstract
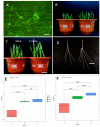
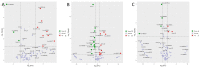
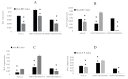
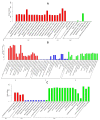
The interactions between plants and microorganisms, which are widely present in the microbial-dominated rhizosphere, have been studied. This association is highly beneficial to the organisms involved, as plants benefit soil microorganisms by providing them with metabolites, while microorganisms promote plant growth and development by promoting nutrient uptake and/or protecting the plant from biotic and abiotic stresses. Piriformospora indica, an endophytic fungus of Sebacinales, colonizes the roots of a wide range of host plants and establishes various benefits for the plants. In this work, an interaction between barley and the P. indica was established to elucidate microRNA (miRNA)-based regulatory changes in miRNA profiles and gene expression that occurred during the symbiosis. Growth promotion and vigorous root development were confirmed in barley colonized by P. indica. The genome-wide expression profile analysis of miRNAs in barley root showed that 7,798,928, 6,418,039 and 7,136,192 clean reads were obtained from the libraries of mock, 3 dai and 7 dai roots, respectively. Sequencing of the barley genome yielded in 81 novel miRNA and 450 differently expressed genes (DEGs). Additionally, 11, 24, 6 differentially expressed microRNAs (DEMs) in barley were found in the three comparison groups, including 3 dai vs. mock, 7 dai vs. mock and 7 dai vs. 3 dai, respectively. The predicted target genes of these miRNAs are mainly involved in transcription, cell division, auxin signal perception and transduction, photosynthesis and hormone stimulus. Transcriptome analysis of P. indica identified 667 and 594 differentially expressed genes (DEG) at 3 dai and 7 dai. Annotation and GO (Gene Ontology) analysis indicated that the DEGs with the greatest changes were concentrated in oxidoreductase activity, ion transmembrane transporter activity. It implies that reprogramming of fundamental miRNA and gene expression occurs both in barley and P. indica. Analysis of global changes in miRNA profiles of barley colonized with P. indica revealed that several putative endogenous barley miRNAs expressed upon colonization belonging to known micro RNA families involved in growth and developmental regulation.
Keywords: Piriformospora indica; RNA-seq; barley; miRNA; reprogramming; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Deshmukh S., Huckelhoven R., Schäfer P., Imani J., Sharma M., Weiss M., Waller F., Kogel K.-H. The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc. Natl. Acad. Sci. USA. 2006;103:18450–18457. doi: 10.1073/pnas.0605697103. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

