The Botrytis cinerea Gene Expression Browser
- PMID: 36675905
- PMCID: PMC9861337
- DOI: 10.3390/jof9010084
The Botrytis cinerea Gene Expression Browser
Abstract
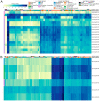
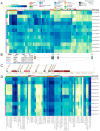
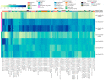
For comprehensive gene expression analyses of the phytopathogenic fungus Botrytis cinerea, which infects a number of plant taxa and is a cause of substantial agricultural losses worldwide, we developed BEB, a web-based B. cinerea gene Expression Browser. This computationally inexpensive web-based application and its associated database contain manually curated RNA-Seq data for B. cinerea. BEB enables expression analyses of genes of interest under different culture conditions by providing publication-ready heatmaps depicting transcript levels, without requiring advanced computational skills. BEB also provides details of each experiment and user-defined gene expression clustering and visualization options. If needed, tables of gene expression values can be downloaded for further exploration, including, for instance, the determination of differentially expressed genes. The BEB implementation is based on open-source computational technologies that can be deployed for other organisms. In this case, the new implementation will be limited only by the number of transcriptomic experiments that are incorporated into the platform. To demonstrate the usability and value of BEB, we analyzed gene expression patterns across different conditions, with a focus on secondary metabolite gene clusters, chromosome-wide gene expression, previously described virulence factors, and reference genes, providing the first comprehensive expression overview of these groups of genes in this relevant fungal phytopathogen. We expect this tool to be broadly useful in B. cinerea research, providing a basis for comparative transcriptomics and candidate gene identification for functional assays.
Keywords: BEB; Botrytis Expression Browser; Botrytis cinerea; RNA-Seq; phytopathogen; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

