In Vitro Fermentation of Pleurotus eryngii Mushrooms by Human Fecal Microbiota: Metataxonomic Analysis and Metabolomic Profiling of Fermentation Products
- PMID: 36675949
- PMCID: PMC9865116
- DOI: 10.3390/jof9010128
In Vitro Fermentation of Pleurotus eryngii Mushrooms by Human Fecal Microbiota: Metataxonomic Analysis and Metabolomic Profiling of Fermentation Products
Abstract
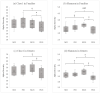
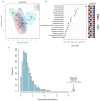
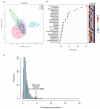
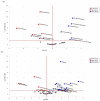
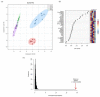
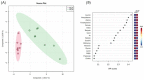
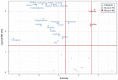
Edible mushrooms contain biologically active compounds with antioxidant, antimicrobial, immunomodulatory and anticancer properties. The link between their anticancer and immunomodulatory properties with their possible prebiotic activity on gut micro-organisms has been the subject of intense research over the last decade. Lyophilized Pleurotus eryngii (PE) mushrooms, selected due to their strong lactogenic effect and anti-genotoxic, immunomodulatory properties, underwent in vitro static batch fermentation for 24 h by fecal microbiota from eight elderly apparently healthy volunteers (>65 years old). The fermentation-induced changes in fecal microbiota communities were examined using Next Generation Sequencing of the hypervariable regions of the 16S rRNA gene. Primary processing and analysis were conducted using the Ion Reporter Suite. Changes in the global metabolic profile were assessed by 1H NMR spectroscopy, and metabolites were assigned by 2D NMR spectroscopy and the MetaboMiner platform. PLS-DA analysis of both metataxonomic and metabolomic data showed a significant cluster separation of PE fermented samples relative to controls. DEseq2 analysis showed that the abundance of families such as Lactobacillaceae and Bifidobacteriaceae were increased in PE samples. Accordingly, in metabolomics, more than twenty metabolites including SCFAs, essential amino acids, and neurotransmitters discriminate PE samples from the respective controls, further validating the metataxonomic findings.
Keywords: Pleurotus eryngii mushrooms; gut microbiota; in vitro static batch fermentation; metabolomics; metataxonomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Finimundy T.C., Gambato G., Fontana R., Camassola M., Salvador M., Moura S., Hess J., Henriques J.A.P., Dillon A.J.P., Roesch-Ely M. Aqueous extracts of Lentinula edodes and Pleurotus sajor-caju exhibit high antioxidant capability and promising in vitro antitumor activity. Nutr. Res. 2013;33:76–84. doi: 10.1016/j.nutres.2012.11.005. - DOI - PubMed
-
- Baraza L.D., Neser W., Jackson K.C., Fredrick J.B., Dennis O., Wairimu K.R., Keya A.O., Heydenreich M. Antimicrobial Coumarins from the Oyster Culinary-Medicinal Mushroom, Pleurotus ostreatus (Agaricomycetes), from Kenya. Int. J. Med. Mushrooms. 2016;18:905–913. doi: 10.1615/IntJMedMushrooms.v18.i10.60. - DOI - PubMed
-
- Boulaka A., Christodoulou P., Vlassopoulou M., Koutrotsios G., Bekiaris G., Zervakis G.I., Mitsou E.K., Saxami G., Kyriacou A., Zervou M., et al. Genoprotective Properties and Metabolites of β-Glucan-Rich Edible Mushrooms Following Their In Vitro Fermentation by Human Faecal Microbiota. Molecules. 2020;25:3554. doi: 10.3390/molecules25153554. - DOI - PMC - PubMed
-
- Arora S., Tandon S. Mushroom Extracts Induce Human Colon Cancer Cell (COLO-205) Death by Triggering the Mitochondrial Apoptosis Pathway and Go/G1-Phase Cell Cycle Arrest. Arch. Iran. Med. 2015;18:284–295. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

