Genetic Predisposition to Hepatocellular Carcinoma
- PMID: 36676960
- PMCID: PMC9864136
- DOI: 10.3390/metabo13010035
Genetic Predisposition to Hepatocellular Carcinoma
Abstract
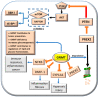
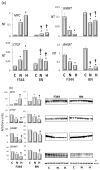
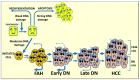
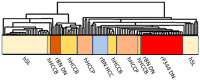
Liver preneoplastic and neoplastic lesions of the genetically susceptible F344 and resistant BN rats cluster, respectively, with human HCC with better (HCCB) and poorer prognosis (HCCP); therefore, they represent a valid model to study the molecular alterations determining the genetic predisposition to HCC and the response to therapy. The ubiquitin-mediated proteolysis of ERK-inhibitor DUSP1, which characterizes HCC progression, favors the unrestrained ERK activity. DUSP1 represents a valuable prognostic marker, and ERK, CKS1, or SKP2 are potential therapeutic targets for human HCC. In DN (dysplastic nodule) and HCC of F344 rats and human HCCP, DUSP1 downregulation and ERK1/2 overexpression sustain SKP2-CKS1 activity through FOXM1, the expression of which is associated with a susceptible phenotype. SAM-methyl-transferase reactions and SAM/SAH ratio are regulated by GNMT. In addition, GNMT binds to CYP1A, PARP1, and NFKB and PREX2 gene promoters. MYBL2 upregulation deregulates cell cycle and induces the progression of premalignant and malignant liver. During HCC progression, the MYBL2 transcription factor positively correlates with cells proliferation and microvessel density, while it is negatively correlated to apoptosis. Hierarchical supervised analysis, regarding 6132 genes common to human and rat liver, showed a gene expression pattern common to normal liver of both strains and BN nodules, and a second pattern is observed in F344 nodules and HCC of both strains. Comparative genetics studies showed that DNs of BN rats cluster with human HCCB, while F344 DNs and HCCs cluster with HCCP.
Keywords: BN rats; CYP1A; DUSP1; F344 rats; FOXM1; GNMT; MYBL2 transcription factor; PARP1 NFKB and PREX2 genes; hepatocellular carcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

