Optimization of Transposon Mutagenesis Methods in Pseudomonas antarctica
- PMID: 36677410
- PMCID: PMC9864612
- DOI: 10.3390/microorganisms11010118
Optimization of Transposon Mutagenesis Methods in Pseudomonas antarctica
Abstract
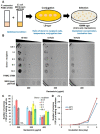
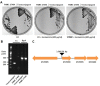
Pseudomonas is a widespread genus in various host and environmental niches. Pseudomonas exists even in extremely cold environments such as Antarctica. Pseudomonas antarctica is a psychrophilic bacterium isolated from Antarctica. P. antarctica is also known to produce antimicrobial substances. Although P. antarctica can provide insight into how bacteria have adapted to low temperatures and has significant potential for developing novel antimicrobial substances, progress in genetic and molecular studies has not been achieved. Transposon mutagenesis is a useful tool to screen genes of interest in bacteria. Therefore, we attempted for the first time in P. antarctica to generate transposon insertion mutants using the transfer of a conjugational plasmid encoding a transposon. To increase the yield of transposon insertion mutants, we optimized the methods, in terms of temperature for conjugation, the ratio of donor and recipient during conjugation, and the concentration of antibiotics. Here, we describe the optimized methods to successfully generate transposon insertion mutants in P. antarctica.
Keywords: Pseudomonas; Pseudomonas antarctica; transposon; transposon insertion mutant library.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Reddy G.S., Matsumoto G.I., Schumann P., Stackebrandt E., Shivaji S. Psychrophilic pseudomonads from Antarctica: Pseudomonas antarctica sp. nov., Pseudomonas meridiana sp. nov. and Pseudomonas proteolytica sp. nov. Int. J. Syst. Evol. Microbiol. 2004;54:713–719. doi: 10.1099/ijs.0.02827-0. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

