Application of Cloning-Free Genome Engineering to Escherichia coli
- PMID: 36677507
- PMCID: PMC9866961
- DOI: 10.3390/microorganisms11010215
Application of Cloning-Free Genome Engineering to Escherichia coli
Abstract
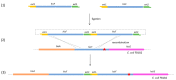
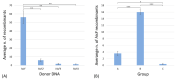
The propagation of foreign DNA in Escherichia coli is central to molecular biology. Recent advances have dramatically expanded the ability to engineer (bacterial) cells; however, most of these techniques remain time-consuming. The aim of the present work was to explore the possibility to use the cloning-free genome editing (CFGE) approach, proposed by Döhlemann and coworkers (2016), for E. coli genetics, and to deepen the knowledge about the homologous recombination mechanism. The E. coli auxotrophic mutant strains FB182 (hisF892) and FB181 (hisI903) were transformed with the circularized wild-type E. coli (i) hisF gene and hisF gene fragments of decreasing length, and (ii) hisIE gene, respectively. His+ clones were selected based on their ability to grow in the absence of histidine, and their hisF/hisIE gene sequences were characterized. CFGE method allowed the recombination of wild-type his genes (or fragments of them) within the mutated chromosomal copy, with a different recombination frequency based on the fragment length, and the generation of clones with a variable number of in tandem his genes copies. Data obtained pave the way to further evolutionary studies concerning the homologous recombination mechanism and the fate of in tandem duplicated genes.
Keywords: evolutionary mechanisms; genetic engineering; histidine biosynthesis; homologous recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

