Integration of Epidemiological and Genomic Data to Investigate H5N1 HPAI Outbreaks in Northern Italy in 2021-2022
- PMID: 36678449
- PMCID: PMC9865711
- DOI: 10.3390/pathogens12010100
Integration of Epidemiological and Genomic Data to Investigate H5N1 HPAI Outbreaks in Northern Italy in 2021-2022
Abstract
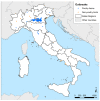
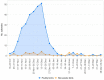
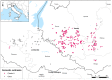
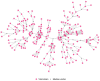
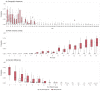
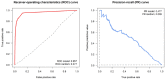
Between October 2021 and April 2022, 317 outbreaks caused by highly pathogenic avian influenza (HPAI) H5N1 viruses were notified in poultry farms in the northeastern Italian regions. The complete genomes of 214 strains were used to estimate the genetic network based on the similarity of the viruses. An exponential random graph model (ERGM) was used to assess the effect of 'at-risk contacts', 'same owners', 'in-bound/out-bound risk windows overlap', 'genetic differences', 'geographic distances', 'same species', and 'poultry company' on the probability of observing a link within the genetic network, which can be interpreted as the potential propagation of the epidemic via lateral spread or a common source of infection. The variables 'same poultry company' (Est. = 0.548, C.I. = [0.179; 0.918]) and 'risk windows overlap' (Est. = 0.339, C.I. = [0.309; 0.368]) were associated with a higher probability of link formation, while the 'genetic differences' (Est. = -0.563, C.I. = [-0.640; -0.486]) and 'geographic distances' (Est. = -0.058, C.I. = [-0.078; -0.038]) indicated a reduced probability. The integration of epidemiological data with genomic analyses allows us to monitor the epidemic evolution and helps to explain the dynamics of lateral spreads casting light on the potential diffusion routes. The 2021-2022 epidemic stresses the need to further strengthen the biosecurity measures, and to encourage the reorganization of the poultry production sector to minimize the impact of future epidemics.
Keywords: ERGM; H5N1; HPAI; Italy; contact tracing; epidemiological investigation; genetic network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Multiple Introductions of Reassorted Highly Pathogenic Avian Influenza H5Nx Viruses Clade 2.3.4.4b Causing Outbreaks in Wild Birds and Poultry in The Netherlands, 2020-2021.Microbiol Spectr. 2022 Apr 27;10(2):e0249921. doi: 10.1128/spectrum.02499-21. Epub 2022 Mar 14. Microbiol Spectr. 2022. PMID: 35286149 Free PMC article.
-
Risk Prediction of Three Different Subtypes of Highly Pathogenic Avian Influenza Outbreaks in Poultry Farms: Based on Spatial Characteristics of Infected Premises in South Korea.Front Vet Sci. 2022 May 31;9:897763. doi: 10.3389/fvets.2022.897763. eCollection 2022. Front Vet Sci. 2022. PMID: 35711796 Free PMC article.
-
Spatiotemporal analysis of highly pathogenic avian influenza (H5N1) outbreaks in poultry in Egypt (2006 to 2017).BMC Vet Res. 2022 May 12;18(1):174. doi: 10.1186/s12917-022-03273-w. BMC Vet Res. 2022. PMID: 35550145 Free PMC article.
-
The potential spread of highly pathogenic avian influenza virus via dynamic contacts between poultry premises in Great Britain.BMC Vet Res. 2011 Oct 13;7:59. doi: 10.1186/1746-6148-7-59. BMC Vet Res. 2011. PMID: 21995783 Free PMC article.
-
[Highly pathogenic avian influenza in poultry (fowl plague); implications for human health].Bull Acad Natl Med. 2005 Nov;189(8):1817-26. Bull Acad Natl Med. 2005. PMID: 16737105 Review. French.
Cited by
-
Study of the Interface between Wild Bird Populations and Poultry and Their Potential Role in the Spread of Avian Influenza.Microorganisms. 2023 Oct 21;11(10):2601. doi: 10.3390/microorganisms11102601. Microorganisms. 2023. PMID: 37894259 Free PMC article.
-
Stakeholders' Perceptions of Biosecurity Implementation in Italian Poultry Farms.Animals (Basel). 2023 Oct 18;13(20):3246. doi: 10.3390/ani13203246. Animals (Basel). 2023. PMID: 37893970 Free PMC article.
References
-
- Mulatti P., Ferrè N., Marangon S. Spatial Distribution of 2000-2007 Low Pathogenicity Avian Influenza Epidemics in Northern Italy. In: Majumdar S., Brenner F., Huffman J., McLean R., Panah A., Pietrobon P., Keeler S., Shive S., editors. Pandemic Influenza Viruses: Science, Surveillance and Public Health. Pennsylvania Academy of Science; Easton, PA, USA: 2011.
-
- Galbraith C.A., Jones T., Kirby J., Mundkur T. In: A Review of Migratory Bird Flyways and Priorities for Management. UNEP/CMS Secretariat, editor. Secretariat of the Convention on the Conservation of Migratory Species of Wild Animals; Bonn, Germany: 2014.
-
- Mulatti P., Fusaro A., Scolamacchia F., Zecchin B., Azzolini A., Zamperin G., Terregino C., Cunial G., Monne I., Marangon S. Integration of Genetic and Epidemiological Data to Infer H5N8 HPAI Virus Transmission Dynamics during the 2016–2017 Epidemic in Italy. Sci. Rep. 2018;8:18037. doi: 10.1038/s41598-018-36892-1. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

