A Synthetic Polymicrobial Community Biofilm Model Demonstrates Spatial Partitioning, Tolerance to Antimicrobial Treatment, Reduced Metabolism, and Small Colony Variants Typical of Chronic Wound Biofilms
- PMID: 36678466
- PMCID: PMC9862141
- DOI: 10.3390/pathogens12010118
A Synthetic Polymicrobial Community Biofilm Model Demonstrates Spatial Partitioning, Tolerance to Antimicrobial Treatment, Reduced Metabolism, and Small Colony Variants Typical of Chronic Wound Biofilms
Abstract
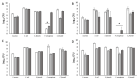
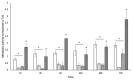
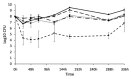
Understanding chronic wound infection is key for successful treatment and requires accurate laboratory models. We describe a modified biofilm flow device that effectively mimics the chronic wound environment, including simulated wound fluid, a collagen-based 3D biofilm matrix, and a five-species mixture of clinically relevant bacteria (Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, Enterococcus faecalis, and Citrobacter freundii). Mixed biofilms were cultured for between 3 and 14 days with consistent numbers of bacteria that exhibited reduced metabolic activity, which increased with a high dose of glucose. S. aureus was recovered from biofilms as a small colony variant, but as a normal colony variant if P. aeruginosa was excluded from the system. Bacteria within the biofilm did not co-aggregate but formed discrete, species-specific clusters. Biofilms demonstrated differential tolerance to the topical antimicrobials Neosporin and HOCl, consistent with protection due to the biofilm lifestyle. The characteristics exhibited within this model match those of real-world wound biofilms, reflecting the clinical scenario and yielding a powerful in vitro tool that is versatile, inexpensive, and pivotal for understanding chronic wound infection.
Keywords: antimicrobial; polymicrobial; wound infection.
Conflict of interest statement
L.I.R. is a consultant at Briotech Incorporated. The authors declare no other conflict of interest.
Figures

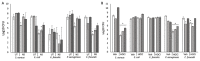



References
-
- Fazli M., Bjarnsholt T., Kirketerp-Møller K., Jørgensen B., Andersen A.S., Krogfelt K.A., Givskov M., Tolker-Nielsen T. Nonrandom Distribution of Pseudomonas Aeruginosa and Staphylococcus Aureus in Chronic Wounds. J. Clin. Microbiol. 2009;47:4084–4089. doi: 10.1128/JCM.01395-09. - DOI - PMC - PubMed
-
- Hinchliffe R.J., Valk G.D., Apelqvist J., Armstrong D.G., Bakker K., Game F.L., Hartemann-Heurtier A., Löndahl M., Price P.E., van Houtum W.H., et al. A Systematic Review of the Effectiveness of Interventions to Enhance the Healing of Chronic Ulcers of the Foot in Diabetes. Diabetes Metab. Res. Rev. 2008;24:S119–S144. doi: 10.1002/dmrr.825. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

