Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain
- PMID: 36678572
- PMCID: PMC9865020
- DOI: 10.3390/ph16010075
Identification of Novel Small Molecule Ligands for JAK2 Pseudokinase Domain
Abstract
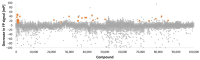
Hyperactive mutation V617F in the JAK2 regulatory pseudokinase domain (JH2) is prevalent in patients with myeloproliferative neoplasms. Here, we identified novel small molecules that target JH2 of JAK2 V617F and characterized binding via biochemical and structural approaches. Screening of 107,600 small molecules resulted in identification of 55 binders to the ATP-binding pocket of recombinant JAK2 JH2 V617F protein at a low hit rate of 0.05%, which indicates unique structural characteristics of the JAK2 JH2 ATP-binding pocket. Selected hits and structural analogs were further assessed for binding to JH2 and JH1 (kinase) domains of JAK family members (JAK1-3, TYK2) and for effects on MPN model cell viability. Crystal structures were determined with JAK2 JH2 wild-type and V617F. The JH2-selective binders were identified in diaminotriazole, diaminotriazine, and phenylpyrazolo-pyrimidone chemical entities, but they showed low-affinity, and no inhibition of MPN cells was detected, while compounds binding to both JAK2 JH1 and JH2 domains inhibited MPN cell viability. X-ray crystal structures of protein-ligand complexes indicated generally similar binding modes between the ligands and V617F or wild-type JAK2. Ligands of JAK2 JH2 V617F are applicable as probes in JAK-STAT research, and SAR optimization combined with structural insights may yield higher-affinity inhibitors with biological activity.
Keywords: JAK inhibitor; JAK2 V617F; cytokine signaling; myeloproliferative neoplasm; pseudokinase.
Conflict of interest statement
This study was supported by grants from Academy of Finland, Sigrid Jusélius Foundation, Jane and Aatos Erkko Foundation, Finnish Cancer Foundation, Tampere Tuberculosis Foundation, and Pirkanmaa hospital district competitive research funding. T.H. reports grant from Novo Nordisk Foundation. P.S. is currently an employee of Surrozen Inc. M.P. is currently an employee of MedEngine Oy. S.R.H. is a co-founder and scientific advisory board member of Ajax Therapeutics, Inc. O.S. reports lecture fees from Pfizer, Abbvie and Novartis, is a member of scientific advisory board of Ajax Therapeutics and chair of scientific advisory board of Finnish hematology registry and biobank. O.S. holds patents on JAK kinases, US Patent no. 5,728,536, US patent no. 8,841,078, AU 2011214254, CAN 2789186, and EPO 11741946.5, and reports royalties or licenses from St Jude Children’s Research Hospital and stock option for Ajax Therapeutics. A.T.V, S.L., J.L. and N.N. declare no competing interests.
Figures




References
-
- George Abraham B., Raivola J., Virtanen A., Silvennoinen O. Janus Kinase. In: Offermanns S., Rosenthal W., editors. Encyclopedia of Molecular Pharmacology. Springer International Publishing; Cham, Switzerland: 2021. pp. 893–902.
-
- Min X., Ungureanu D., Maxwell S., Hammarén H., Thibault S., Hillert E.K., Ayres M., Greenfield B., Eksterowicz J., Gabel C., et al. Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2) J. Biol. Chem. 2015;290:27261–27270. doi: 10.1074/jbc.M115.672048. - DOI - PMC - PubMed
-
- Ungureanu D., Wu J., Pekkala T., Niranjan Y., Young C., Jensen O.N., Xu C.F., Neubert T.A., Skoda R.C., Hubbard S.R., et al. The Pseudokinase Domain of JAK2 Is a Dual-Specificity Protein Kinase That Negatively Regulates Cytokine Signaling. Nat. Struct. Mol. Biol. 2011;18:971–976. doi: 10.1038/nsmb.2099. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

