Structural Refinement by Direct Mapping Reveals Assembly Inconsistencies near Hi-C Junctions
- PMID: 36679033
- PMCID: PMC9861903
- DOI: 10.3390/plants12020320
Structural Refinement by Direct Mapping Reveals Assembly Inconsistencies near Hi-C Junctions
Abstract
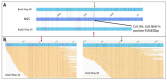
High-throughput chromosome conformation capture (Hi-C) is widely used for scaffolding in de novo assembly because it produces highly contiguous genomes, but its indirect statistical approach can introduce connection errors. We employed optical mapping (Bionano Genomics) as an orthogonal scaffolding technology to assess the structural solidity of Hi-C reconstructed scaffolds. Optical maps were used to assess the correctness of five de novo genome assemblies based on long-read sequencing for contig generation and Hi-C for scaffolding. Hundreds of inconsistencies were found between the reconstructions generated using the Hi-C and optical mapping approaches. Manual inspection, exploiting raw long-read sequencing data and optical maps, confirmed that several of these conflicts were derived from Hi-C joining errors. Such misjoins were widespread, involved the connection of both small and large contigs, and even overlapped annotated genes. We conclude that the integration of optical mapping data after, not before, Hi-C-based scaffolding, improves the quality of the assembly and limits reconstruction errors by highlighting misjoins that can then be subjected to further investigation.
Keywords: Hi-C; assembly refinement; de novo genome assembly; optical mapping.
Conflict of interest statement
Author E.C. is an employee of Genartis srl. Authors M.R. and M.D. are partners of Genartis srl. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Lee J.Y., Kong M., Oh J., Lim J.S., Chung S.H., Kim J.M., Kim J.S., Kim K.H., Yoo J.C., Kwak W. Comparative Evaluation of Nanopore Polishing Tools for Microbial Genome Assembly and Polishing Strategies for Downstream Analysis. Sci. Rep. 2021;11:20740. doi: 10.1038/s41598-021-00178-w. - DOI - PMC - PubMed
-
- Xue Z., Chen-guang L., Shi-hui Y., Xia W., Feng-wu B., Zhuo W. Benchmarking of long-read sequencing, assemblers and polishers for yeast genome. Brief. Bioinform. 2022;23:bbac146. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

