Unglycosylated Soluble SARS-CoV-2 Receptor Binding Domain (RBD) Produced in E. coli Combined with the Army Liposomal Formulation Containing QS21 (ALFQ) Elicits Neutralizing Antibodies against Mismatched Variants
- PMID: 36679887
- PMCID: PMC9864931
- DOI: 10.3390/vaccines11010042
Unglycosylated Soluble SARS-CoV-2 Receptor Binding Domain (RBD) Produced in E. coli Combined with the Army Liposomal Formulation Containing QS21 (ALFQ) Elicits Neutralizing Antibodies against Mismatched Variants
Abstract
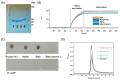
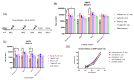
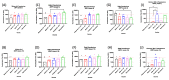
The emergence of novel potentially pandemic pathogens necessitates the rapid manufacture and deployment of effective, stable, and locally manufacturable vaccines on a global scale. In this study, the ability of the Escherichia coli expression system to produce the receptor binding domain (RBD) of the SARS-CoV-2 spike protein was evaluated. The RBD of the original Wuhan-Hu1 variant and of the Alpha and Beta variants of concern (VoC) were expressed in E. coli, and their biochemical and immunological profiles were compared to RBD produced in mammalian cells. The E. coli-produced RBD variants recapitulated the structural character of mammalian-expressed RBD and bound to human angiotensin converting enzyme (ACE2) receptor and a panel of neutralizing SARS-CoV-2 monoclonal antibodies. A pilot vaccination in mice with bacterial RBDs formulated with a novel liposomal adjuvant, Army Liposomal Formulation containing QS21 (ALFQ), induced polyclonal antibodies that inhibited RBD association to ACE2 in vitro and potently neutralized homologous and heterologous SARS-CoV-2 pseudoviruses. Although all vaccines induced neutralization of the non-vaccine Delta variant, only the Beta RBD vaccine produced in E. coli and mammalian cells effectively neutralized the Omicron BA.1 pseudovirus. These outcomes warrant further exploration of E. coli as an expression platform for non-glycosylated, soluble immunogens for future rapid response to emerging pandemic pathogens.
Keywords: E. coli; RBD; SARS-CoV-2; recombinant; vaccine.
Conflict of interest statement
GM has a patent the ALFQ adjuvant. No other author has any conflict of interest. The funders had no role in the design the study or its interpretation.
Figures



Similar articles
-
A Glycosylated RBD Protein Induces Enhanced Neutralizing Antibodies against Omicron and Other Variants with Improved Protection against SARS-CoV-2 Infection.J Virol. 2022 Sep 14;96(17):e0011822. doi: 10.1128/jvi.00118-22. Epub 2022 Aug 16. J Virol. 2022. PMID: 35972290 Free PMC article.
-
Functional Expression of the Recombinant Spike Receptor Binding Domain of SARS-CoV-2 Omicron in the Periplasm of Escherichia coli.Bioengineering (Basel). 2022 Nov 10;9(11):670. doi: 10.3390/bioengineering9110670. Bioengineering (Basel). 2022. PMID: 36354581 Free PMC article.
-
Non-glycosylated SARS-CoV-2 RBD elicited a robust neutralizing antibody response in mice.J Immunol Methods. 2022 Jul;506:113279. doi: 10.1016/j.jim.2022.113279. Epub 2022 May 6. J Immunol Methods. 2022. PMID: 35533747 Free PMC article.
-
RBD-mRNA vaccine induces broadly neutralizing antibodies against Omicron and multiple other variants and protects mice from SARS-CoV-2 challenge.Transl Res. 2022 Oct;248:11-21. doi: 10.1016/j.trsl.2022.04.007. Epub 2022 Apr 28. Transl Res. 2022. PMID: 35489692 Free PMC article.
-
Utilization of Receptor-Binding Domain of SARS-CoV-2 Spike Protein Expressed in Escherichia coli for the Development of Neutralizing Antibody Assay.Mol Biotechnol. 2023 Apr;65(4):598-611. doi: 10.1007/s12033-022-00563-4. Epub 2022 Sep 14. Mol Biotechnol. 2023. PMID: 36103078 Free PMC article.
Cited by
-
A Clinical Update on SARS-CoV-2: Pathology and Development of Potential Inhibitors.Curr Issues Mol Biol. 2023 Jan 4;45(1):400-433. doi: 10.3390/cimb45010028. Curr Issues Mol Biol. 2023. PMID: 36661514 Free PMC article. Review.
-
Production and Immunogenicity Assessment of LTp50: An Escherichia coli-Made Chimeric Antigen Targeting S1- and S2-Epitopes from the SARS-CoV-2/BA.5 Spike Protein.Pharmaceuticals (Basel). 2024 Feb 27;17(3):302. doi: 10.3390/ph17030302. Pharmaceuticals (Basel). 2024. PMID: 38543088 Free PMC article.
-
Oral administration of a recombinant modified RBD antigen of SARS-CoV-2 as a possible immunostimulant for the care of COVID-19.Microb Cell Fact. 2024 Feb 6;23(1):41. doi: 10.1186/s12934-024-02320-5. Microb Cell Fact. 2024. PMID: 38321489 Free PMC article.
-
Antisera Produced Using an E. coli-Expressed SARS-CoV-2 RBD and Complemented with a Minimal Dose of Mammalian-Cell-Expressed S1 Subunit of the Spike Protein Exhibits Improved Neutralization.Int J Mol Sci. 2023 Jun 24;24(13):10583. doi: 10.3390/ijms241310583. Int J Mol Sci. 2023. PMID: 37445760 Free PMC article.
-
A bioengineered pseudovirus nanoparticle displaying SARS-CoV 2 RBD fully protects mice from mortality and weight loss caused by SARS-CoV 2 challenge.Biotechnol J. 2023 Oct;18(10):e2300130. doi: 10.1002/biot.202300130. Epub 2023 Jun 23. Biotechnol J. 2023. PMID: 37300425 Free PMC article.
References
-
- Hemida M.G., A Perera R., Wang P., A Alhammadi M., Siu L.Y., Li M., Poon L.L., Saif L., Alnaeem A., Peiris M. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Eurosurveillance. 2013;18:20659. doi: 10.2807/1560-7917.ES2013.18.50.20659. - DOI - PubMed
-
- Dhama K., Patel S.K., Sharun K., Pathak M., Tiwari R., Yatoo M.I., Malik Y.S., Sah R., Rabaan A.A., Panwar P.K., et al. SARS-CoV-2 jumping the species barrier: Zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med. Infect. Dis. 2020;37:101830. doi: 10.1016/j.tmaid.2020.101830. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

