Microcalorimetry: A Novel Application to Measure In Vitro Phage Susceptibility of Staphylococcus aureus in Human Serum
- PMID: 36680055
- PMCID: PMC9865112
- DOI: 10.3390/v15010014
Microcalorimetry: A Novel Application to Measure In Vitro Phage Susceptibility of Staphylococcus aureus in Human Serum
Abstract
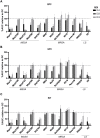
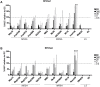
Infections involving antibiotic resistant Staphylococcus aureus (S. aureus) represent a major challenge to successful treatment. Further, although bacteriophages (phages) could be an alternative to antibiotics, there exists a lack of correlation in phage susceptibility results between conventional in vitro and in vivo assays. This discrepancy may hinder the potential implementation of bacteriophage therapy. In this study, the susceptibility of twelve S. aureus strains to three commercial phage cocktails and two single phages was assessed. These S. aureus strains (including ten clinical isolates, five of which were methicillin-resistant) were compared using four assays: the spot test, efficiency of plating (EOP), the optical density assay (all in culture media) and microcalorimetry in human serum. In the spot test, EOP and optical density assay, all cocktails and single phages lysed both methicillin susceptible and methicillin resistant S. aureus strains. However, there was an absence of phage-mediated lysis in high concentrations of human serum as measured using microcalorimetry. As this microcalorimetry-based assay more closely resembles in vivo conditions, we propose that microcalorimetry could be included as a useful addition to conventional assays, thereby facilitating more accurate predictions of the in vivo susceptibility of S. aureus to phages during phage selection for therapeutic purposes.
Keywords: MRSA; Staphylococcus aureus; bacteriophage therapy; human serum; microcalorimetry; susceptibility testing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cassini A., Högberg L.D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G.S., Colomb-Cotinat M., Kretzschmar M.E., Devleesschauwer B., Cecchini M., et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. - DOI - PMC - PubMed
-
- Delattre R., Seurat J., Haddad F., Nguyen T.-T., Gaborieau B., Kane R., Dufour N., Ricard J.-D., Guedj J., Debarbieux L. Title combination of in vivo phage therapy data with in silico model highlights key parameters for treatment efficacy. Cell Rep. 2022;39:110825. doi: 10.1016/j.celrep.2022.110825. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

