Phage Encounters Recorded in CRISPR Arrays in the Genus Oenococcus
- PMID: 36680056
- PMCID: PMC9867325
- DOI: 10.3390/v15010015
Phage Encounters Recorded in CRISPR Arrays in the Genus Oenococcus
Abstract
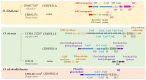
The Oenococcus genus comprises four recognized species, and members have been found in different types of beverages, including wine, kefir, cider and kombucha. In this work, we implemented two complementary strategies to assess whether oenococcal hosts of different species and habitats were connected through their bacteriophages. First, we investigated the diversity of CRISPR-Cas systems using a genome-mining approach, and CRISPR-endowed strains were identified in three species. A census of the spacers from the four identified CRISPR-Cas loci showed that each spacer space was mostly dominated by species-specific sequences. Yet, we characterized a limited records of potentially recent and also ancient infections between O. kitaharae and O. sicerae and phages of O. oeni, suggesting that some related phages have interacted in diverse ways with their Oenococcus hosts over evolutionary time. Second, phage-host interaction analyses were performed experimentally with a diversified panel of phages and strains. None of the tested phages could infect strains across the species barrier. Yet, some infections occurred between phages and hosts from distinct beverages in the O. oeni species.
Keywords: CRISPR; Oenococcus; bacteriophage; fermented beverage; host spectra; spacers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Phage-host interactions analysis of newly characterized Oenococcus oeni bacteriophages: Implications for malolactic fermentation in wine.Int J Food Microbiol. 2017 Apr 4;246:12-19. doi: 10.1016/j.ijfoodmicro.2017.01.020. Epub 2017 Jan 31. Int J Food Microbiol. 2017. PMID: 28189899
-
Phylogenomic Analysis of Oenococcus oeni Reveals Specific Domestication of Strains to Cider and Wines.Genome Biol Evol. 2015 May 14;7(6):1506-18. doi: 10.1093/gbe/evv084. Genome Biol Evol. 2015. PMID: 25977455 Free PMC article.
-
Natural diversity of CRISPR spacers of Thermus: evidence of local spacer acquisition and global spacer exchange.Philos Trans R Soc Lond B Biol Sci. 2019 May 13;374(1772):20180092. doi: 10.1098/rstb.2018.0092. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 30905291 Free PMC article.
-
Distribution of Oenococcus oeni populations in natural habitats.Appl Microbiol Biotechnol. 2019 Apr;103(7):2937-2945. doi: 10.1007/s00253-019-09689-z. Epub 2019 Feb 20. Appl Microbiol Biotechnol. 2019. PMID: 30788540 Free PMC article. Review.
-
Exploring the diversity of bacteriophage specific to Oenococcus oeni and Lactobacillus spp and their role in wine production.Appl Microbiol Biotechnol. 2021 Dec;105(23):8575-8592. doi: 10.1007/s00253-021-11509-2. Epub 2021 Oct 25. Appl Microbiol Biotechnol. 2021. PMID: 34694447 Review.
Cited by
-
Oenococcus oeni allows the increase of antihypertensive and antioxidant activities in apple cider.Heliyon. 2023 Jun 1;9(6):e16806. doi: 10.1016/j.heliyon.2023.e16806. eCollection 2023 Jun. Heliyon. 2023. PMID: 37332959 Free PMC article.
References
-
- International Organisation of Vine and Wine [OIV] Annual Report. International Organisation of Vine and Wine; Paris, France: 2021.
-
- Vitulo N., Lemos W.J.F., Jr., Calgaro M., Confalone M., Felis G.E., Zapparoli G., Nardi T. Bark and grape microbiome of Vitis vinifera: Influence of geographic patterns and agronomic management on bacterial diversity. Front. Microbiol. 2019;9:3203. doi: 10.3389/fmicb.2018.03203. - DOI - PMC - PubMed
-
- Fournier P., Pellan L., Barroso-Bergad D., Bohanc D.A., Candresse T., Delmotte F., Dufour M.-C., Lauvergeat V., Le Marrec C., Marais A., et al. The functional microbiome of grapevine throughout plant evolutionary history and lifetime. Adv. Ecol. Res. 2022. in press .
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

